- EN - English
- CN - 中文
Microscopic Detection of DNA Synthesis in Early Mitosis at Repetitive lacO Sequences in Human Cells
人细胞重复 lacO 序列中早期有丝分裂中 DNA 合成的显微镜检测
发布: 2022年09月05日第12卷第17期 DOI: 10.21769/BioProtoc.4504 浏览次数: 2669
评审: Gal HaimovichDavid PaulVaibhav B. Shah
Abstract
In the human cell cycle, complete replication of DNA is a fundamental process for the maintenance of genome integrity. Replication stress interfering with the progression of replication forks causes difficult-to-replicate regions to remain under-replicated until the onset of mitosis. In early mitosis, a homology-directed repair DNA synthesis, called mitotic DNA synthesis (MiDAS), is triggered to complete DNA replication. Here, we present a method to detect MiDAS in human U2OS 40-2-6 cells, in which repetitive lacO sequences integrated into the human chromosome evoke replication stress and concomitant incomplete replication of the lacO array. Immunostaining of BrdU and LacI proteins is applied for visualization of DNA synthesis in early mitosis and the lacO array, respectively. This protocol has been established to easily detect MiDAS at specific loci using only common immunostaining methods and may be optimized for the investigation of other difficult-to-replicate regions marked with site-specific binding proteins.
Keywords: DNA replication (基因复制)Background
A basic principle of the cell cycle is to ensure that mitosis follows the completion of DNA replication. During S phase, various replication stresses, such as DNA lesions, conflicts with transcription, and tightly bound protein complexes, interfere with DNA replication and cause a genomic instability related to various genetic diseases (Muñoz and Méndez, 2017). To prevent or minimize the deleterious consequences of replication stress, cells have evolved DNA damage response mechanisms, including DNA repair pathways (Yoshida and Fujita, 2021). Stalled replication forks activate checkpoint pathways that halt the cell cycle progression (Lemmens and Lindqvist, 2019; Mocanu and Chan, 2021). However, it has been revealed that upon mild replication stress that modestly slows down the replication fork, cells can enter the mitosis with some under-replicated DNA (Lukas et al., 2011; Bertolin et al., 2020; Lezaja and Altmeyer, 2021; Mocanu and Chan, 2021). Mild replication stress causes incomplete replication without activating G2/M checkpoint, especially at difficult-to-replicate regions such as chromosomal fragile sites, centromeres, and telomeres (Sarlós et al., 2017; Özer and Hickson, 2018; Lokanga et al., 2021). Such regions with under-replicated DNA activate mitotic DNA synthesis (MiDAS) in early mitosis to complete DNA replication, whereas some regions remain incomplete and will be resolved in the subsequent cell cycle (Minocherhomji et al., 2015; Özer and Hickson, 2018; Bertolin et al., 2020; Lezaja and Altmeyer, 2021).
MiDAS is a kind of homology-directed repair DNA synthesis that involves repair of stalled replication intermediates and POLD3 (an accessory subunit of polymerase δ)-dependent conservative DNA synthesis, analogous to break-induced replication (Kockler et al., 2021; Epum and Haber, 2022). Basic protocols for detection of MiDAS were originally developed by Hickson and colleagues (Minocherhomji et al., 2015; Bhowmick et al., 2016; Garribba et al., 2018). Briefly, cells are treated with low dose (0.4 μM) aphidicolin and RO-3306. Aphidicolin, a DNA polymerase inhibitor, slows down DNA replication and induces under-replication, while RO-3306, an inhibitor of cyclin-dependent kinase 1, arrests cells at G2 phase. Cells are then released into mitosis in the presence of a nucleoside analog ethynyl deoxyuridine (EdU) to label newly synthesized DNA. Following click reaction, MiDAS is visualized as EdU foci on prophase chromatin by fluorescence microscopy.
Here, we describe a detailed method for detection of MiDAS at under-replicated regions evoked by repetitive lacO sequences in human U2OS 40-2-6 cells (Figure 1). U2OS 40-2-6 cell, a U2OS-derived cell line carrying lacO array and estrogen receptor (ERT2)–fused LacI gene, was generated to investigate DNA damage response to replication stress induced by tight DNA–protein complexes on the human chromosome (Ishimoto et al., 2021). Treatment with 4-hydroxytamoxifen (4-OHT) induces rapid formation of lacO–LacI complexes that interfere with DNA replication. Notably, we also found that even in the absence of LacI, repetitive lacO sequences are intrinsically difficult to replicate, remaining under-replicated until mitosis and consequently triggering MiDAS. In the method presented here, nascent DNA is labeled by another nucleoside analog bromodeoxyuridine (BrdU), and lacO array is visualized by immunostaining of LacI proteins bound to the array. In our ER–LacI induction system, replication stress is induced at the specific chromosomal locus using the sequence-specific DNA-binding proteins. MiDAS at the specific locus can therefore be easily detected using common immunostaining methods, without fluorescent in situ hybridization (FISH) used in the protocol developed by Hickson and colleagues. This protocol may be optimized for other repetitive fragile sites where MiDAS functions, such as telomeres, centromeres, and ribosomal DNA arrays.
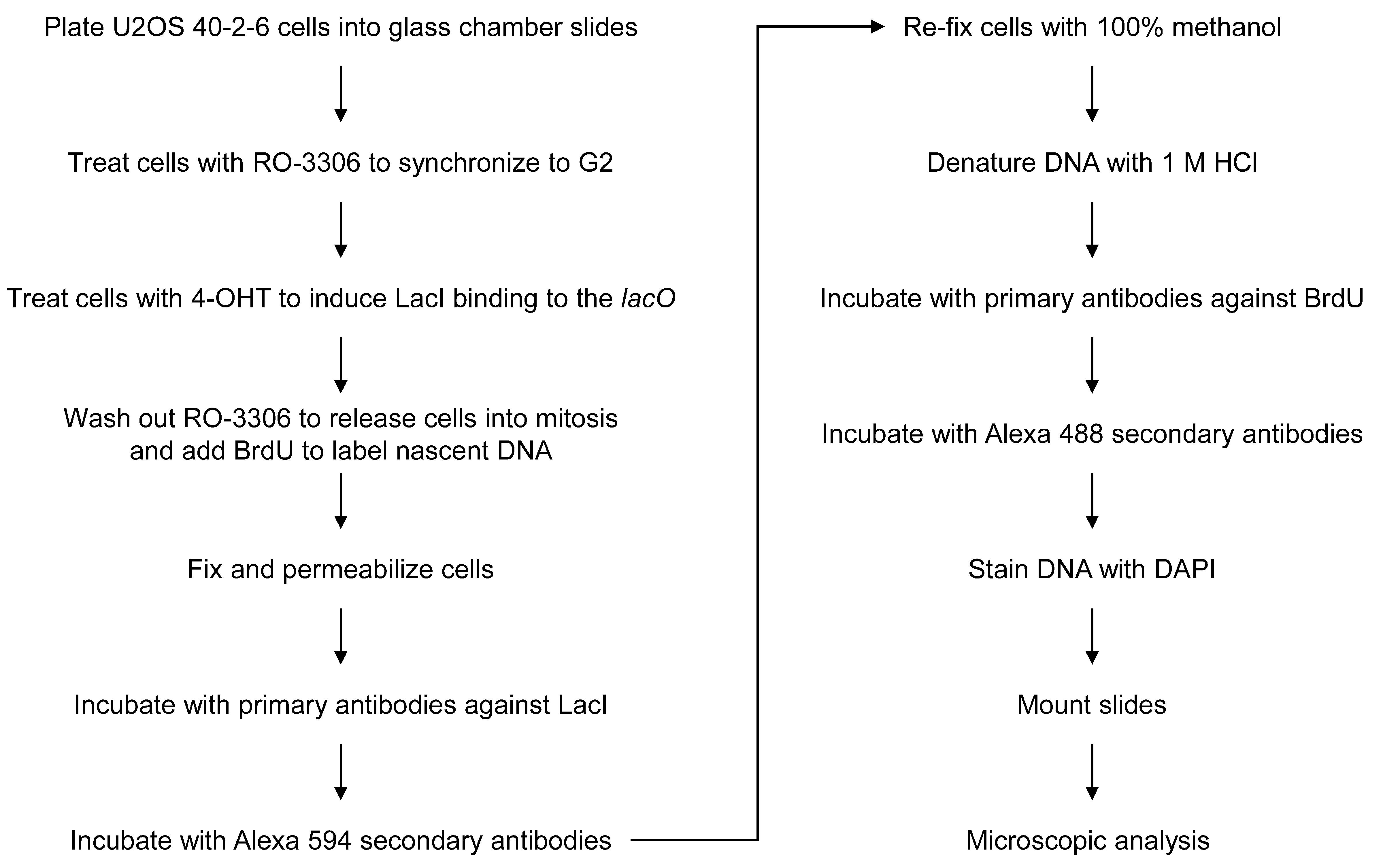
Figure 1. Flowchart of the assay for detection of mitotic DNA synthesis at the lacO array.
Materials and Reagents
Lab-Tek II, 4-well chamber glass slides (Thermo Fisher, Nunc, catalog number: 154526)
Chamber removal tool (included with chamber slides)
Glass Coplin jar, 5 slides type (AsOne, catalog number: 4-567-01)
Syringe filter unit, PES 0.22 μm (Merk Millipore, catalog number: SLGPR33RS)
Syringe filter unit, PVDF 0.45 μm (Merk Millipore, catalog number: SLHVR33RS)
Rectangular dish (Eiken Chemical, catalog number: AW2000)
Cotton swab
Paper towel (Nippon Paper Crecia, catalog number: 37105)
Glass coverslips, 24 × 60 mm (Matsunami Glass, catalog number: C024601)
Nail polish (clear topcoat)
Human cell line U2OS 40-2-6 (Ishimoto et al., 2021), a U2OS-derived cell line carrying lacO array and ERT2-LacI gene generated from U2OS 2-6-3 cells (Janicki et al., 2004). U2OS 40-2-6 cells, and the original U2OS 2-6-3 cells, contain an approximately 200-copy array of 256 lacO repeats (approximately 50,000 copies of lacO sequences in total). The array is stably integrated into the chromosomal locus 1p36 in chromosome 1.
Dulbecco’s modified Eagle’s medium (D-MEM) (high glucose with L-glutamine) (e.g., Wako, catalog number: 043-30085)
Fetal bovine serum (FBS) (e.g., Nichirei, catalog number: 175012)
RO-3306 (e.g., Sigma-Aldrich, catalog number: SML0569)
4-Hydroxytamoxifen (4-OHT) (e.g., Abcam, catalog number: ab141943)
Ethanol (e.g., Nacalai Tesque, catalog number: 14713-53)
10× phosphate-buffered saline (PBS) (e.g., Wako, catalog number: 048-29805), diluted to 1×
Bromodeoxyuridine (BrdU) (e.g., Sigma-Aldrich, catalog number: B5002)
Saline (e.g., Otsuka Normal Saline, Otsuka Pharmaceutical)
PIPES (e.g., Dojindo, catalog number: 347-02224)
0.1 M EGTA-2Na solution (e.g., Nacalai Tesque, catalog number: 37346-05)
Triton X-100 (e.g., Wako, catalog number: 169-21105)
MgCl2 (e.g., Wako, Catalog number: 133-00165)
37% formaldehyde solution (e.g., Nacalai Tesque, catalog number: 16223-55)
Rabbit anti-LacI antibody, homemade (Ishimoto et al., 2021)
Thimerosal (e.g., Nacalai Tesque, Catalog number: 21624-74), dissolved with water to 10% (w/v)
Alexa 594 donkey anti-Rabbit IgG (Molecular Probes, catalog number: A21207)
Methanol (e.g., Nacalai Tesque, catalog number: 21915-93)
HCl (e.g., Wako, catalog number: 080-01066), diluted to 1 M
Mouse anti-BrdU antibody, clone: 3D4 (BD Pharmingen, catalog number: 555627)
Alexa 488 goat anti-Mouse IgG (Molecular Probes, catalog number: A11029)
4’, 6-diamidino-2-phenylindole (DAPI) (e.g., Wako, catalog number: 043-18804)
Fluoro-KEEPER antifade reagent (Nacalai Tesque, catalog number: 12593-64)
RO-3306 stock solution (see Recipes)
4-OHT stock solution (see Recipes)
BrdU stock solution (see Recipes)
PTEMF buffer (see Recipes)
FBS for antibody working solution (see Recipes)
Antibody working solutions (see Recipes)
DAPI stock solution (see Recipes)
Note: In our lab, we routinely use DAPI, Alexa 488, and Alexa 594 for triple staining of DNA and two interest proteins. Other combinations of fluorophores may be possible.
Equipment
Cell culture incubator (ASTEC, model: SCA-165D)
Water aspirator (e.g., FLINN scientific, model: AP1136)
Widefield fluorescence microscope equipped with a cooled CCD camera, an LED illumination source, and filters to image Alexa 488, Alexa 594, and DAPI (e.g., Keyence, BZ-X700). Similar conventional fluorescence microscopes would be applicable.
40× objective lens [e.g., Nikon, CFI (chromatic aberration free infinity) Plan Apo λ 40×/0.95 NA]. Higher magnification objectives, such as a 63× lens, may also be applicable.
Software
Microscope operating software (e.g., Keyence, the BZ-X viewer software)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yoshida, K., Ishimoto, R. and Fujita, M. (2022). Microscopic Detection of DNA Synthesis in Early Mitosis at Repetitive lacO Sequences in Human Cells. Bio-protocol 12(17): e4504. DOI: 10.21769/BioProtoc.4504.
- Ishimoto, R., Tsuzuki, Y., Matsumura, T., Kurashige, S., Enokitani, K., Narimatsu, K., Higa, M., Sugimoto, N., Yoshida, K. and Fujita, M. (2021). SLX4-XPF mediates DNA damage responses to replication stress induced by DNA-protein interactions. J Cell Biol 220(1): e202003148.
分类
癌症生物学 > 基因组不稳定性及突变 > 细胞生物学试验 > DNA结构和改变
癌症生物学 > 通用技术 > 细胞生物学试验 > 细胞周期
细胞生物学 > 细胞成像 > 荧光
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link