- EN - English
- CN - 中文
Cost Effective Method for gDNA Isolation from the Cecal Content and High Yield Procedure for RNA Isolation from the Colonic Tissue of Mice
从小鼠盲肠内容物中分离gDNA的经济有效方法和从小鼠结肠组织中分离RNA的高产率方法
发布: 2022年08月05日第12卷第15期 DOI: 10.21769/BioProtoc.4484 浏览次数: 2187
评审: Alba BlesaTimo A LehtiChao Jiang
Abstract
Microbiome studies are quickly gaining momentum. Since most of the resident microbes (consisting of bacteria, fungi, and viruses) are difficult to culture, sequencing the microbial genome is the method of choice to characterize them. It is therefore important to have efficient methodology for gDNA isolation of gut microbes. Mouse models are widely used to understand human disease etiology while avoiding human ethics-related complications. However, the widely used kit-based methods are costly, and sometimes yields (in terms of quality and quantity) are sub-optimal. To overcome this problem, we developed a straightforward, standardized DNA isolation procedure from mouse cecal content for further microbiome-related studies. The reagents we used to standardize the procedure are readily available even in a not-so-well-equipped laboratory, and the reagents are not expensive. The yield and quality of the DNA are also better than those obtained by the readily available kit-based methods.
Additionally, we modified the kit-based method of RNA isolation from the colon tissue sample of the mouse for better yield. Churning the tissue with liquid nitrogen at the beginning of the procedure improves RNA quality and quantity.
Graphical abstract:
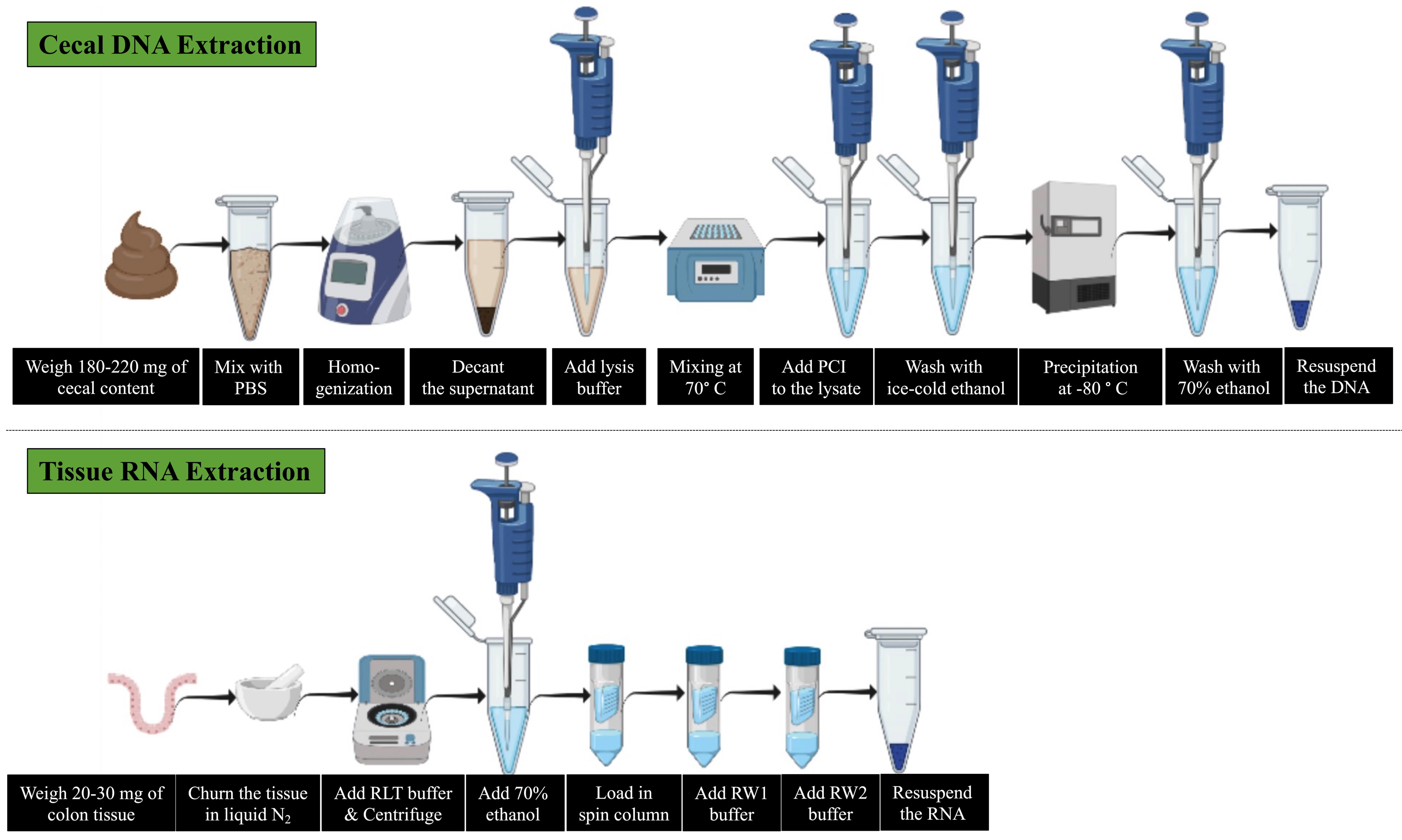
Background
Efficient extraction of high-quality (concentration: 800–2,200 ng/μL) and high molecular weight (>50 kb) genomic DNA and tissue RNA from the limited amount of available samples is the key challenge for downstream applications like next-generation DNA sequencing (NGS) or transcriptomics studies, among others. The expanding research on human diseases related to gut microbiota and associated gut health resulted in a plethora of methodologies used to profile the gut microbial composition and associated inflammatory status of the gut tissue.
Each step in the downstream data analysis process is subject to technical variation depending on the protocol or materials used for DNA or RNA isolation. Researchers have found striking differences in the results obtained from different kit-based methods within the same laboratory (Ferrand et al., 2014; Videnska et al., 2019; Gryp et al., 2020). Therefore, standardizing the protocols is of utmost priority for researchers to obtain consistent, good quality, and robust representative data.
The diversity of the gut microbes in gut ecosystems can be categorized based on their morphological, structural, biochemical, and genetic properties. Gut microbes also have notable differences regarding cell wall structure and cell membranes, which enclose their cytoplasm and genomic contents. Harsh sample treatments can affect DNA quality, while a mild process may cause partial lysis of the microbial community (Bag et al., 2016). Moreover, the unknown composition of the lysis buffer of kit-based methods presents different biases, drastically changing the results obtained from the same sample (Brooks et al., 2015). Therefore, it is important to optimize cell lysis methods to obtain genomic DNA from all groups of microbes present in the gut.
We developed a very efficient, cost-effective cell lysis method. Reagents used to standardize the process are readily available, even in a comparatively less equipped laboratory, and the reagents are not too expensive. The yield and quality of the DNA are also better than those obtained by the readily available kit-based methods.
In an RNA isolation procedure from gut tissue, it is essential to standardize the amount of tissue required to obtain good quality RNA. Proper tissue homogenization is another crucial step for a high yield of RNA. To get high quality and quantity of RNA, we standardized the needed amount of colonic tissue and the liquid nitrogen-based homogenization process.
These methodologies were published in a recent article (Pradhan et al., 2022).
Materials and Reagents
For gDNA isolation
Filter MAXIPENSETM, Low retention, clear, sterile tips 1,000 μL (Tarsons, catalog number: 527106), 200 μL (Tarsons, catalog number: 527104), and 10 μL (Tarsons, catalog number: 527100)
SpinwinTM Micro Centrifuge clear, sterile tubes 1.5 mL (Tarsons, catalog number: 500010) and 2 mL (Tarsons, catalog number: 500020)
Dissection box
Spatula
Tissue paper
Kimwipes
Nuclease-free water (Promega, catalog number: P1193)
1× sterile PBS (HiMedia, catalog number: TL1022)
Phenol-chloroform-isoamyl alcohol (24:24:1) (Sigma-Aldrich, catalog number: 516276)
Ice-cold absolute ethanol (Sigma-Aldrich, catalog number: 459836)
RNase A (Sigma-Aldrich, catalog number: R6513)
Tris-base
HCl
EDTA
NaCl
SDS
Milli-Q® ultra-pure water
70% ethanol (1,000 mL) (see Recipes)
Lysis buffer (100 mL), pH 7.2 (see Recipes)
For tissue RNA isolation
Filter MAXIPENSETM, Low retention, clear, sterile tips 1,000 μL (Tarsons, catalog number: 527106), 200 μL (Tarsons, catalog number: 527104), and 10 μL (Tarsons, catalog number: 527100)
SpinwinTM Micro Centrifuge clear, sterile tubes 1.5 mL (Tarsons, catalog number: 500010) and 2 mL (Tarsons, catalog number: 500020)
Dissection box
1 mL syringe
Weighing paper (Fisher Brand, size 4" × 4")
Tissue paper
Kimwipes
Mortar and pestle
RNAlater® (Sigma-Aldrich, catalog number: R0901)
RNaseZAPTM (Sigma-Aldrich, catalog number: R2020)
Nuclease-free water (Promega, catalog number: P1193)
Liquid nitrogen
RNeasy Mini Kit (Qiagen, catalog number: 74104)
DNase I (Qiagen, catalog number: 74106)
RDD buffer (Qiagen, catalog number: 74106)
70% ethanol (1,000 mL) (see Recipes)
Equipment
Pipette (1,000 μL, 200 μL, 20 μL, 2 μL) (Thermo Fisher, Massachusetts, U.S)
-80 °C freezer (Thermo Fisher, Massachusetts, U.S)
CO2 chamber for mice (Thermo Fisher, Massachusetts, U.S)
Vortex (Fisher Biotec, Wembley WA 6904, Australia)
Microcentrifuge (Eppendorf, Hamburg, Germany)
Laminar airflow (Thermo Fisher Scientific, Columbus, OH, USA)
Thermoshaker (Thermo Fisher, Massachusetts, U.S)
Rotospin (Tarsons, Kolkata, India) or any test tube rotator
Nanodrop 2000 machine (Thermo Fisher Scientific, Columbus, OH, USA) or any UV-VIS spectrophotometer (minimum sample volume 0–5 μL)
Qubit4 fluorometer (Invitrogen, California, USA) or any normal fluorometer
Horizontal gel electrophoresis apparatus (California, U.S)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Mukhopadhyay, S. and Aich, P. (2022). Cost Effective Method for gDNA Isolation from the Cecal Content and High Yield Procedure for RNA Isolation from the Colonic Tissue of Mice. Bio-protocol 12(15): e4484. DOI: 10.21769/BioProtoc.4484.
分类
免疫学 > 动物模型 > 小鼠
微生物学 > 微生物-宿主相互作用 > 细菌
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link














