- EN - English
- CN - 中文
Investigation of Transposon DNA Methylation and Copy Number Variation in Plants Using Southern Hybridisation
利用Southern杂交技术研究植物中转座子DNA甲基化和拷贝数变异的情况
发布: 2022年06月05日第12卷第11期 DOI: 10.21769/BioProtoc.4432 浏览次数: 3411
评审: Prashanth N SuravajhalaAnil KumarWenyang LiAnsul LokdarshiAnonymous reviewer(s)
Abstract
Plant genomes are pronouncedly enriched in repeat elements such as transposons. These repeats are epigenetically regulated by DNA methylation. Whole genome high-depth sequencing after bisulfite treatment remains an expensive and laborious method to reliably profile the DNA methylome, especially when considering large genomes such as in crops. Here, we present a simple reproducible Southern hybridisation–based assay to obtain incontrovertible methylation patterns from targeted regions in the rice genome. By employing minor but key modifications, we reliably detected transposon copy number variations over multiple generations. This method can be regarded as a gold standard for validation of epigenetic variations at target loci, and the consequent proliferation of transposons, or segregation in several plant replicates and genotypes.
Graphical abstract:
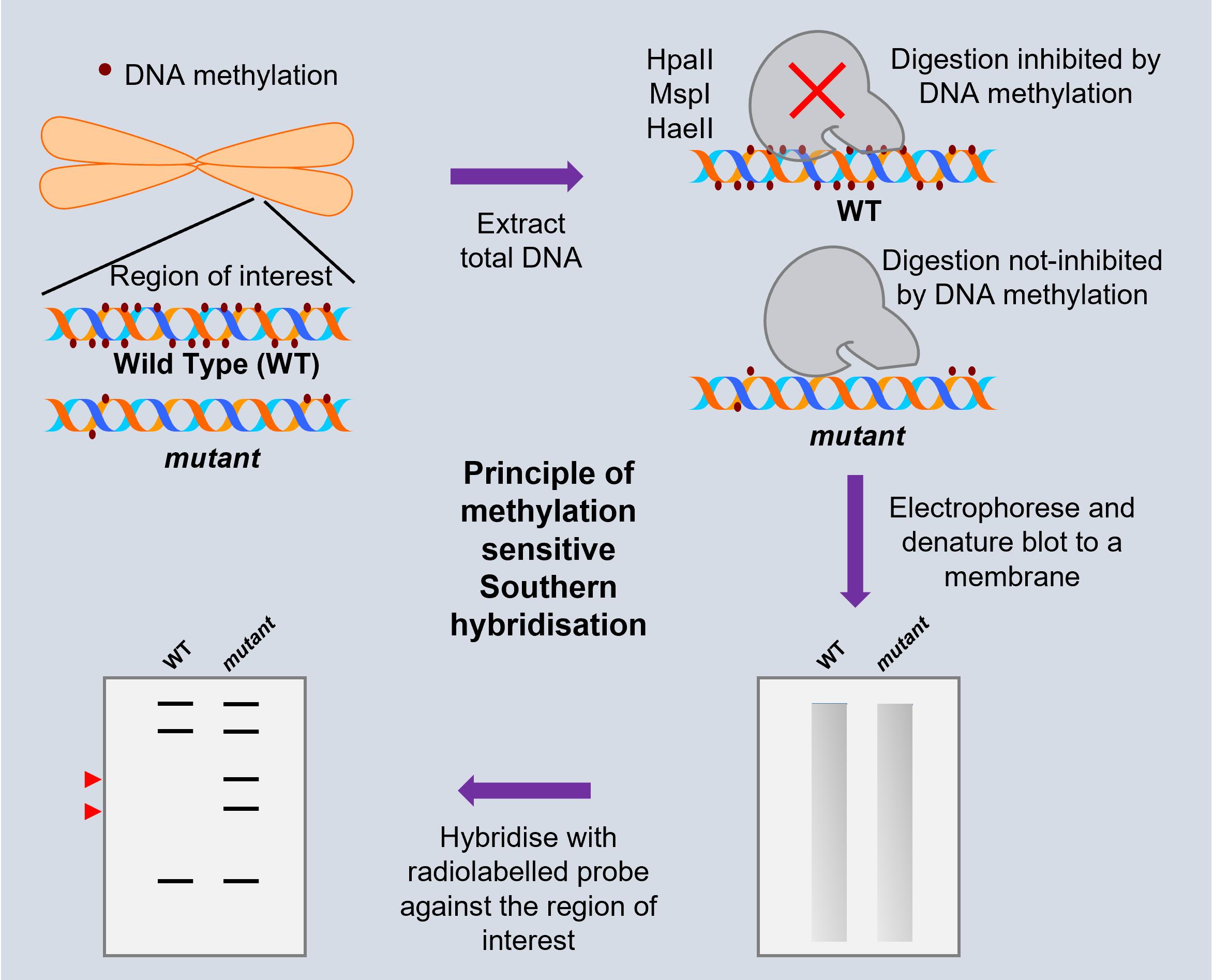
Background
Epigenetics mediated by DNA methylation plays an important role in plant development, regulation of repeats and transposons, immunity, stress tolerance, and transgenerational memory (Zhang et al., 2018). Profiling methylation at targeted loci provides the primary data in delineating the causes and consequences of DNA methylation across genotypes or treatments. This can be investigated by methylation-sensitive enzyme digested DNA PCR (ChoP-PCR), or bisulfite amplicon sequencing (BSAS) (Zhang et al., 2014; Masser et al., 2015). However, these methods have the inherent bias in PCR amplification, and require the usage of primers with very low GC content, which tend to amplify non-specific regions. Hence, additional validation by high throughput methods is necessary for conclusiveness.
Southern hybridisation, on the other hand, involves restriction of the genomic DNA using specific DNA methylation responsive restriction enzyme(s), followed by electrophoresis, blotting, and hybridisation, using targeted regions of interest. This offers several advantages for targeted methylation profiling, a few of which are enumerated below:
Unbiased detection of all the copies of transposons, irrespective of methylation status, truncation, or sequence variants.
DNA methylation among eukaryotes can be distinguished into CG, CHG, and CHH contexts, based on the adjacent nucleotides to cytosines in the genome. Methylation in these contexts is distinctly modulated, and interpreted by different cellular processes. This method offers the ability to distinguish between these different contexts of DNA methylation, by employing restriction enzymes specific to each category.
The hybridised membrane can be stripped and re-probed with multiple probes, which may serve as control regions.
Regions such as rDNA repeats and others, which are recalcitrant for amplification due to presence of a tandem array of repeats, can be profiled for DNA methylation in a reproducible manner.
DNA methylation differences might occur only in a few of the available copies of the specific class of transposons. Such differences that occur in only a few of the identical copies cannot be detected by PCR–based assays. The method described here can reliably detect such variations, as banding patterns differ for every copy of the transposon.
This method can be used to unequivocally prove the proliferation of transposons, upon silencing perturbation, or stress-induced transposon activation. If such an event occurs only in specific tissues or a sub-population of cells, this method can detect such copy number variation.
Since this method characterises the variation in restriction fragments in the genome, the possibility of probing transgene copy variations is an added advantage.
This method can be effectively adapted for any eukaryote. Once considerable quantity and quality of DNA is obtained, the universality of the methylation sensitivity, DNA fragment electrophoresis, and transfer enables probing a region of interest from any source.
Materials and Reagents
Whatman sheets (GE Healthcare, catalog number: 3030-931)
10 mL sterile serological pipette (Biofil, catalog number: GSP211010)
1.5 mL microcentrifuge tubes
Purified total genomic DNA (~5 µg) from each sample
DNA probes designed for the regions of interest
Restriction Enzymes
HpaII (New England Biolabs, catalog number: R0171S)
MspI (New England Biolabs, catalog number: R0106S)
HaeII (New England Biolabs, catalog number: R0107S)
PstI-HF (New England Biolabs, catalog number: R3140S)
EcoRV-HF (New England Biolabs, catalog number: R3195S)
10× CutSmart Buffer (New England Biolabs, catalog number: B7204S)
Ultra-pure water (Invitrogen, catalog number: 10977-023)
Amersham Rediprime II DNA Labeling System (GE Healthcare, catalog number: RPN1633)
Illustra MicroSpin G-50 Columns (GE Healthcare, catalog number: 27533001)
Zeta-Probe® Blotting Membranes (Biorad, catalog number: 162-0165)
UltraPureTM Agarose (Thermo Fisher Scientific, Invitrogen, catalog number: 16500-100)
1 kb DNA ladder (New England Biolabs, catalog number: N3232S)
Sodium hydroxide (Fisher Scientific, catalog number: 27815S)
Sodium chloride (HiMedia, catalog number: MB023-1Kg)
Tris-base (Fisher Scientific, catalog number: 15965-500G)
Hydrochloric acid (Fisher Scientific, catalog number: 29505)
Tri-sodium citrate (Fisher Scientific, catalog number: 14005)
Boric acid (Fisher Scientific, catalog number: 12005)
Ficoll-400 (Sigma, catalog number: F2637)
Bromophenol blue sodium salt (Sigma, catalog number: B5525)
Sodium phosphate dibasic, Molecular weight 141.96 g/mol (Sigma catalog number: 71642)
Double distilled autoclaved water (called “water” unless mentioned otherwise)
Ethidium Bromide (Himedia, catalog number: MB071)
Sodium Dodecyl Sulphate (SDS) (Himedia, catalog number: GRM205)
Ethylene diamine tetra-acetic acid disodium salt (EDTA), Molecular weight 372.24 g/mol (Fisher Scientific, catalog number: 12635)
Ortho-phosphoric acid (Fisher Scientific, catalog number: 29245)
dCTP [α-32P], 10 mCi/mL (Board of Radiation and Isotope Technology, India, catalog number: PLC102)
20× Saline-Sodium citrate (SSC) (see Recipes)
Depurination solution (see Recipes)
Denaturation solution (see Recipes)
Neutralisation solution (see Recipes)
Hybridisation buffer (see Recipes)
Ethidium Bromide staining solution (see Recipes)
2 M sodium phosphate buffer pH 7.2 (see Recipes)
10% SDS (see Recipes)
0.5 M EDTA pH 8.0 (see Recipes)
1 M Tris buffer pH 8.0 (see Recipes)
10× Tris-Borate-EDTA (TBE) buffer (see Recipes)
1× Tris-EDTA (TE) buffer (see Recipes)
8× DNA loading buffer (see Recipes)
Wash buffer 1 (see Recipes)
Wash buffer 2 (see Recipes)
Wash buffer 3 (see Recipes)
Equipment
Water bath (Neolab Instruments, India, model: WB-343)
Microcentrifuge (Eppendorf, model: 5415R)
Thermomixer (Eppendorf, model: Thermomixer comfort)
Chemical fume hood
DNA gel electrophoresis apparatus with accessories (Biorad, Sub-Cell GT Cell)
Hybridisation oven (Thermo Fisher Scientific, Hybaid oven)
Hybridisation cylinders (WHEATON® Safety Coated, Hybridisation bottle, DWK life sciences, catalog number: 805027)
Typhoon Phosphor-Imager scanner system (Amersham Biosciences, GE Healthcare, model: Typhoon TRIO)
Phosphor screen (GE Healthcare, 20×24 cm)
Phosphor exposure cassette (GE Healthcare, 20×24 cm)
Electrophoresis power supply unit (Biorad, PowerPac HC)
Glass trays
Microwave oven
Gel rocker (Bionova India, model: SLMGR100)
Bubble level
UV-gel imaging system (Vilber Gel documentation system)
UV crosslinker (UVP, model: CL1000)
PCR thermal cycler (Biorad, model: S1000)
Radioactive material safe work space with radioactive disposal facility
Software
ImageJ
Typhoon TRIO image acquisition system
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sundar G., V. H. and Shivaprasad, P. V. (2022). Investigation of Transposon DNA Methylation and Copy Number Variation in Plants Using Southern Hybridisation . Bio-protocol 12(11): e4432. DOI: 10.21769/BioProtoc.4432.
分类
植物科学 > 植物分子生物学 > DNA > 基因分型
植物科学 > 植物生物化学
生物化学 > DNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













