- EN - English
- CN - 中文
Labelling of Active Transcription Sites with Argonaute NRDE-3—Image Active Transcription Sites in vivo in Caenorhabditis elegans
用Argonaute NRDE-3标记活性转录位点—图像秀丽隐杆线虫体内的活性转录位点
发布: 2022年06月05日第12卷第11期 DOI: 10.21769/BioProtoc.4427 浏览次数: 2685
评审: Alessandro DidonnaAnand Ramesh PatwardhanAnonymous reviewer(s)
Abstract
Live labelling of active transcription sites is critical to our understanding of transcriptional dynamics. In the most widely used method, RNA sequence MS2 repeats are added to the transcript of interest, on which fluorescently tagged Major Coat Protein binds, and labels transcription sites and transcripts. Here we describe another strategy, using the Argonaute protein NRDE-3, repurposed as an RNA-programmable RNA binding protein. We label active transcription sites in C. elegans embryos and larvae, without editing the gene of interest. NRDE-3 is programmed by feeding nematodes with double-stranded RNA matching the target gene. This method does not require genome editing and is inexpensive and fast to apply to many different genes.
Graphical abstract:
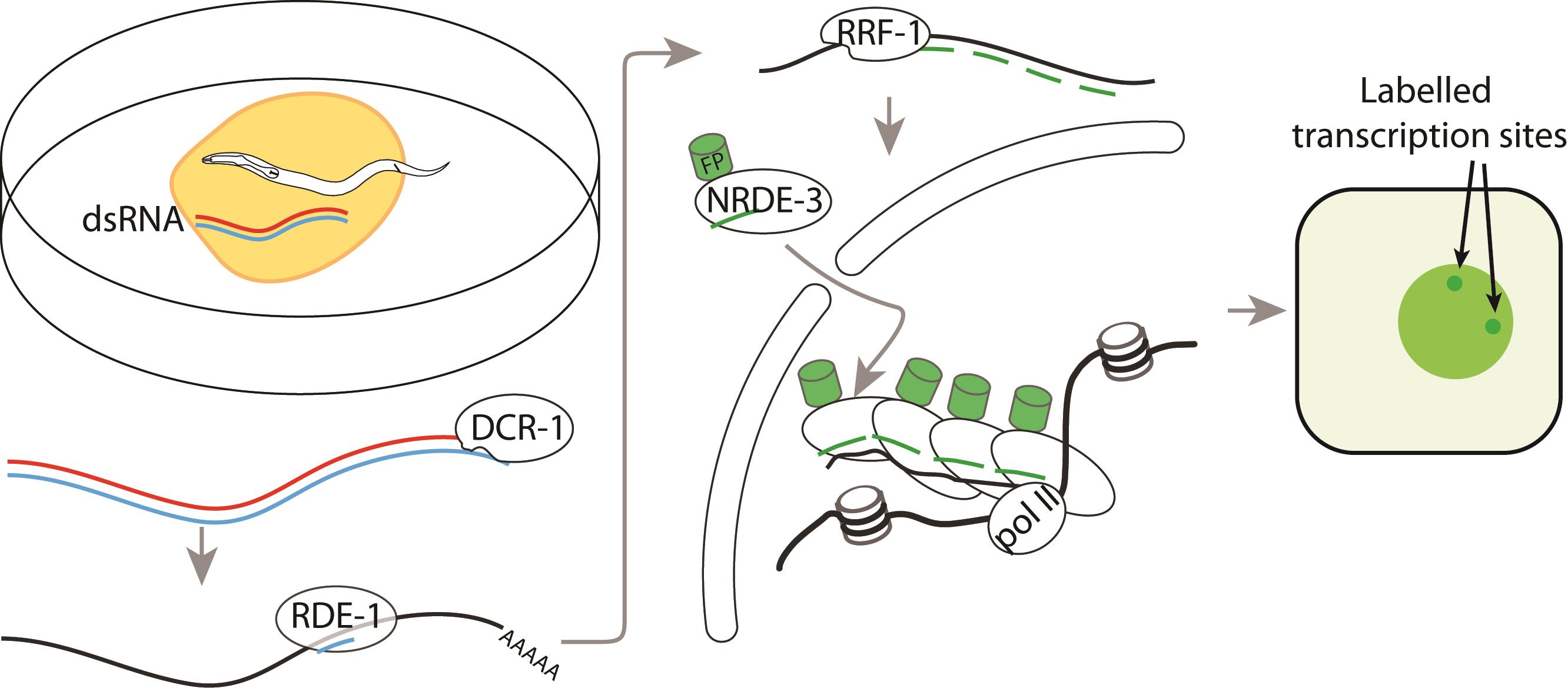
Background
We describe here a protocol to image transcription at transcription sites in C. elegans using an Argonaute protein NRDE-3, based on a method we recently published (Toudji-Zouaz et al., 2021). In C. elegans, double-stranded RNA targeting the gene of interest is uptaken by the systemic RNAi mechanisms and distributed to all cells by the dsRNA transporter SID-1.
The exoRNAi pathway will process dsRNA and use them for recognition of target mRNAs for degradation. This recruits the RNA dependent RNA polymerase RRF-1 to the transcript, which will synthesise triphosphorylated, antisense, secondary small RNAs that are loaded into secondary Argonautes. The only somatic secondary Argonaute, NRDE-3, when unladen, is cytoplasmic. Once loaded with small RNA, NRDE-3 moves to the nucleus and binds to the nascent transcript, where it recruits other proteins to silence transcription.
In a wild-type background, the endogenous RNAi pathway will induce synthesis of secondary small RNAs, and constitutive nuclear localisation of NRDE-3 (Figure 1A, B). To prevent this, we introduce a mutation in eri-1. NRDE-3 is therefore unladen in most tissues (except for the germline, intestine, and early embryo), localises to the cytoplasm, and will move into the nucleus only if the gene of interest is expressed in the cell (Guang et al., 2008) (Figure 1C, D). To prevent downstream transcriptional silencing, we also introduce a mutation in nrde-2 (Guang et al., 2010).
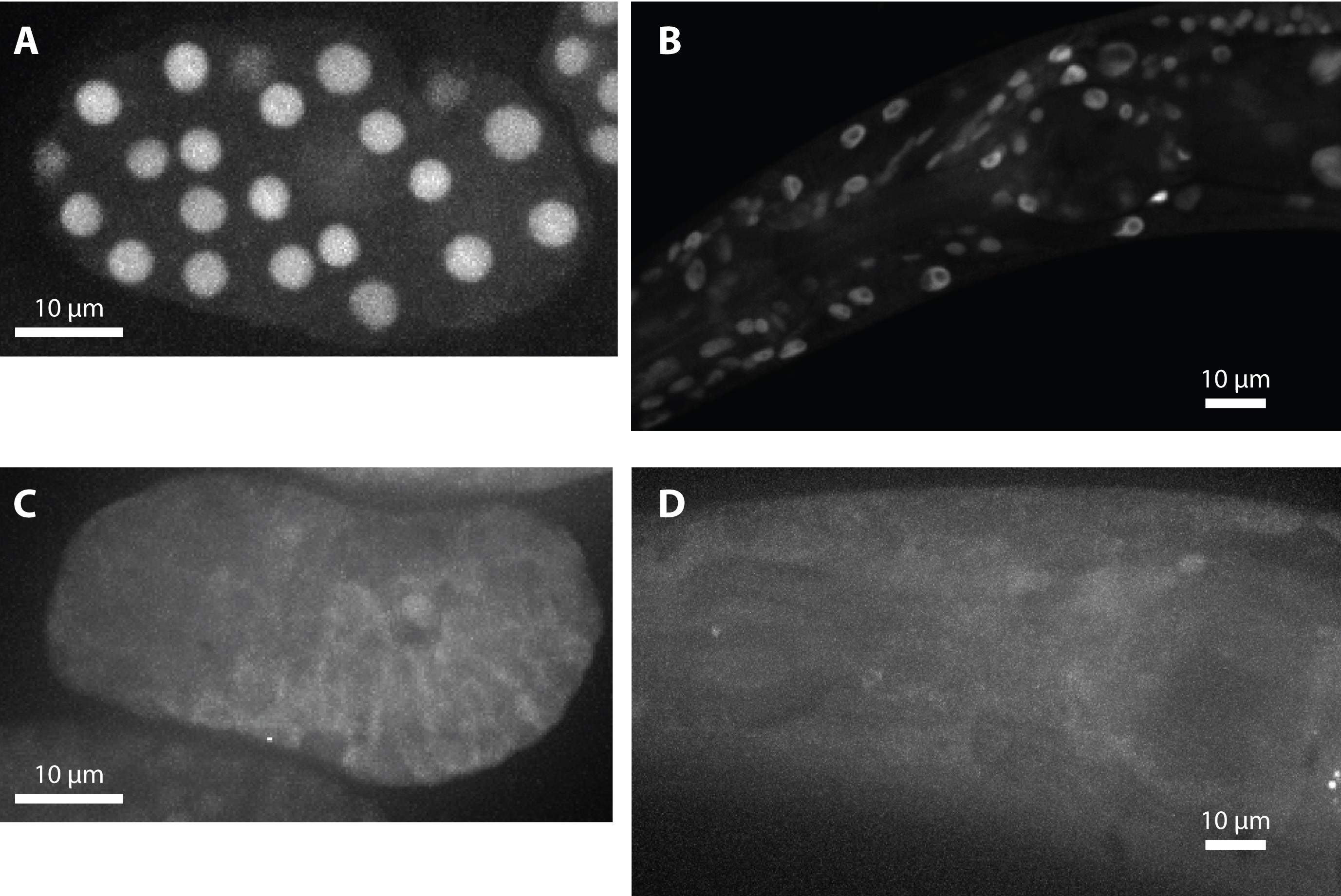
Figure 1. NRDE-3 nuclear localisation is dependent on small RNAs. Nuclear localisation of fluorescently labelled NRDE-3 in the embryo (A) and adult somatic tissues (head) (B) in a eri-1(+) genetic background. Cytoplasmic localisation of NRDE-3 in a eri-1(-) background, in the embryo (C) and adult (D). Note the nuclear localisation in the germline primordium in the embryo (C).
Materials and Reagents
Petri dishes, 60 mm diameter (Falcon, catalog number: 353004)
Microscopy slides (Ghäasel, catalog number: 29-201-307)
Coverslips (Knittel, catalog number: 100037)
Sodium chloride (Roth, CAS: 7647-14-5)
Bacto agar (BD, CAS: 9002-18-0)
Bacto peptone (BD, CAS: 51142-18-8)
5 mg/mL cholesterol in ethanol, filter sterilised (Sigma, CAS: 57-88-5)
KH2PO4 (Roth, CAS 7778-77-0)
K2HPO4 (VWR, CAS: 16788-57-1)
1 M MgSO4 (Roth, CAS: 7487-88-9)
1 M CaCl2 (VWR, CAS: 10035-04-8)
Ampicillin 1,000× (Sigma, CAS: 69-53-4) or carbenicillin 1,000× (Roth, CAS: 4697-36-3) 25 mg/mL (store at -20°C)
IPTG (Sigma, CAS: 367-93-1), 1 M (store at -20°C)
Na2HPO4 (VWR, CAS: 7558-79-4)
Feeding RNAi clones targeting the gene of interest, either from libraries (Ahringer library, Source Bioscience; Vidal library, Horizon discovery, catalog number: RCE1181; stored at -80°C) or homemade
Serotonin (Sigma, catalog number: H7752-5G), stock solution 25 mM in M9, keep at -20°C
0.1 µm diameter polystyrene microspheres (Polysciences, catalog number: 00876-15, 2.5% w/v suspension in M9)
Nematode strains from Caenorhabditis Genetics Center (University of Minnesota): VBS662, VBS663, VBS664, VBS668
Wizard Plus SV miniprep kit (Promega, catalog number: A1460)
LB agar plates with 50 µg/mL ampicillin and 10 µg/mL tetracycline
Liquid LB with 50 µg/mL ampicillin
1 M KPO4 buffer (see Recipes)
M9 buffer (see Recipes)
Equipment
Peristaltic pump (Wheaton Omnispense plus)
Confocal microscope, Spinning disk Roper on Nikon Eclipse Ti, with argon laser for illumination at 515 nm
Incubator (Pol Eko apartura), set at 20°C
Centrifuge (Eppendorf 5804R)
Worm pick, prepared according to (Stiernagle, 2006)
Software
Fiji (https://fiji.sc/)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Barrière, A. and Bertrand, V. (2022). Labelling of Active Transcription Sites with Argonaute NRDE-3—Image Active Transcription Sites in vivo in Caenorhabditis elegans. Bio-protocol 12(11): e4427. DOI: 10.21769/BioProtoc.4427.
分类
细胞生物学 > 细胞成像 > 荧光
生物工程 > 合成生物学
发育生物学 > 细胞生长和命运决定 > 分化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













