- EN - English
- CN - 中文
A Rapid FRET Real-Time PCR Protocol for Simultaneous Quantitative Detection and Discrimination of Human Plasmodium Parasites
一种同时定量检测和鉴别人类疟原虫的快速FRET实时PCR方法
发布: 2022年04月05日第12卷第7期 DOI: 10.21769/BioProtoc.4381 浏览次数: 2372
评审: Alessandro DidonnaEsteban Paredes-OssesDhiman Sankar PalScott McComb
Abstract
Malaria is the most important parasitic disease worldwide, and accurate diagnosis and treatment without delay are essential for reducing morbidity and mortality, especially in P. falciparum malaria. Real-time PCR is highly sensitive and highly specific, therefore an excellent diagnostic tool for laboratory detection and species-specific diagnosis of malaria, especially in non-endemic regions where experience in microscopic malaria diagnostics is limited. In contrast to many other real-time PCR protocols, our new fluorescence resonance energy transfer-based real-time PCR (FRET-qPCR) allows the quantitative and species-specific detection of all human Plasmodium spp. in one run. Species identification is based on single nucleotide polymorphisms (SNPs) within the MalFL probe, detectable by melting curve analysis. A significant advantage of our FRET-qPCR is the short turn-around time of less than two hours, including DNA extraction, which qualifies it for routine diagnostics. Rapid and reliable species-specific malaria diagnosis is important, because treatment is species-dependent. Apart from first-line diagnosis, the quantitative results of our new FRET-qPCR can be helpful in therapy control, to detect early treatment failure.
Graphic abstract:
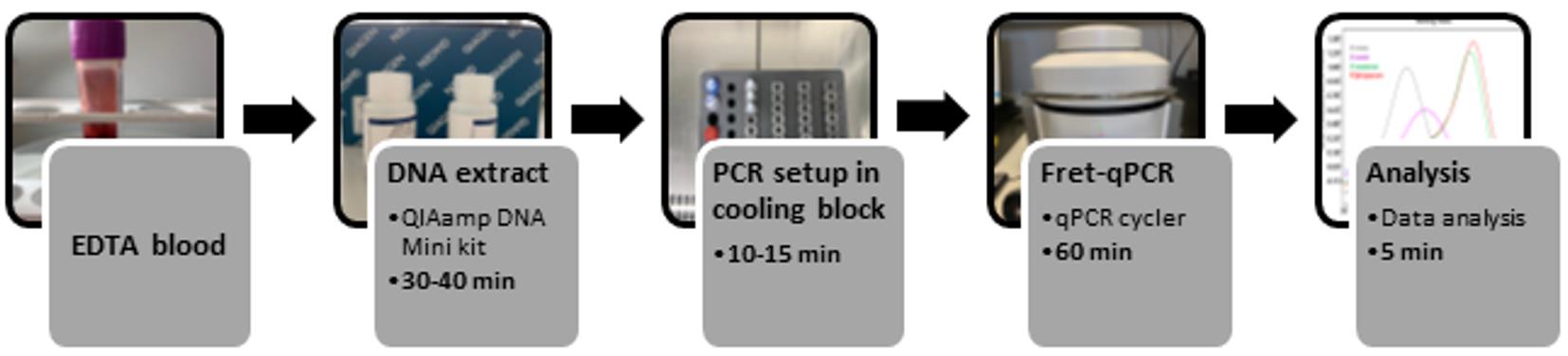
Background
Microscopy of Giemsa stained blood smears is still the gold standard for malaria diagnostics (Yin et al., 2018). However, microscopy requires considerable expertise, particularly at low-level parasitemia, which is often seen in imported malaria in non-endemic countries. Moreover, differentiating the Plasmodium species based on their morphological characteristics can be demanding, especially after chemoprophylaxis or auto-medication (Calderaro et al., 2018). Molecular detection and identification of Plasmodium spp. is a highly accurate and sensitive alternative method for the diagnosis of malaria. Particularly, real-time PCR has the advantage of fast and quantitative results and, compared to nested PCR, the risk of contamination is reduced. However, many published Plasmodium real-time PCR protocols have limitations, e.g., do not differentiate between species (Safeukui et al., 2008; Haanshuus et al., 2019; Farcas et al., 2004), do not detect all species (Perandin et al., 2004;Kim et al., 2014; Veron et al., 2009), or a positive genus-specific PCR has to be followed by two additional multiplex reactions, to obtain a species-specific result (Rougemont et al., 2004).
Hence, we developed and evaluated a new fluorescence resonance energy transfer-based real-time PCR (FRET-qPCR) that allows a quantitative, rapid, sensitive, and species-specific diagnosis of malaria (Schneider et al., 2021). The sequences of the hybridization probes MalFL and MalLC640 match P. falciparum. Fluorescence at 640 nm was generated by FRET, following the annealing of both probes to their adjacent complementary sequences. Species discrimination is based on the presence of single nucleotide polymorphisms (SNPs), reducing the affinity of the MalFLprobe for P. vivax/knowlesi (2 mismatches), P. ovale (1 mismatch), and P. malariae (1 mismatch), and thus lowering the melting temperature (Tm) in the melting curve analysis (Figure 1). The current paper is a step-by-step protocol for the performance of this new FRET-qPCR, focusing on the proper handling and storage of all components. The details described in this protocol are crucial comments to achieve excellent amplification and melting curves, a prerequisite for correct species identification based on single nucleotide polymorphisms (SNPs).
The advantage of requiring only one reaction per patient, and the short turn-around time of less than two hours (including DNA extraction), makes it a valuable diagnostic tool, especially in the absence of experienced microscopists. Moreover, results are objective and reproducible, DNA extraction can be automated, and the use of only one set of primers and probes is a significant advantage in routine clinical applications. One technician, well-trained in molecular methods, will be able to test a large number of patient samples in a relatively short turn-around time.
This protocol can be helpful for the rapid and reliable diagnosis of malaria in individual patients, particularly travelers returning from endemic areas, migrants, and refugees. Another field of application is treatment monitoring, by evaluating the quantitative results, and the crossing point (Cp) values. These allow the determination of the reduction in parasite number, important to detect early treatment failure (Rougemont et al., 2004). The potential of the FRET-qPCRs to detect asymptomatic infections can be useful for screening travelers returning to non-endemic regions. Moreover, it can also be performed in well-equipped malaria reference laboratories in endemic countries.
Materials and Reagents
1.5 mL microcentrifuge tubes, sterile (Sarstedt, catalog number: 72.690.01)
0.5–10 µL extra-long, 100–1,250 µL extra-long, 10–100 µL filtered pipette tips (Biozym, catalog numbers: VT0200, 770600; Eppendorf, catalog number: 30.077.547)
QIAamp® DNA Mini Kit (Qiagen, catalog number: 51304)
Ethanol absolute (Merck, catalog number: 1.00983.2500), do not use denatured alcohol
DNase- and RNase free water (Promega, catalog number: P1193)
LightCycler® Capillaries 20 µL (Roche, catalog number: 4929292001)
LightCycler® Fast Start DNA Master HybProbe (Roche, catalog number: 12239272001)
Primers and hybridization probes (TIB Molbiol GmbH; Table 1)
Primer reconstitution and storage (working stock preparation instructions).
EDTA blood samples
Table 1. Primers and FRET-probes targeting a 157–165 bp fragment of the small subunit 18S rRNA gene, for the detection and simultaneous differentiation of the respective Plasmodium species.
Oligonucleotide Sequence Plasmo 1a
Plasmo 2a
MalFLb
MalLC640b
5′-GTTAAGGGAGTGAAGACGATCAGA-3′
5′-AACCCAAAGACTTTGATTTCTCATAA-3′
5′-CTTTCATCCAACACCTAGTCGGC;
3′label fluorescein
5′-TAGTTTATGGTTAAGATTACGACGGT;
5′label, LC red 640, 3′phosphorylated
aPrimers Plasmo 1 and Plasmo 2 were published byRougemont et al. (2004)
bFRET-probes MalFL and MalLC640 were published bySchneider et al. (2021)
Equipment
0.5–10 µL, 10–100 µL, and 100–1,000 µL micropipettes (Eppendorf, catalog numbers: 3123000020, 3123000047, 3123000063)
ThermoMixer (Eppendorf ThermoMixer C + Smart Block for 1.5 mL tubes; Eppendorf, catalog numbers: 5382000015, EPS360000036)
Microcentrifuge (Thermo ScientificTM, PicoTM 21 Microcentrifuge, catalog number: 75002415)
LightCycler® Instrument 2.0 (Roche, catalog number: 03531414001)
LightCycler® Adapters (Roche, catalog number: 11909312001)
Vortex (Vortex-Genie 2, Sigma-Aldrich, catalog number: Z258423)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Schneider, R., Lamien-Meda, A., Auer, H., Wiedermann-Schmidt, U., Chiodini, P. L. and Walochnik, J. (2022). A Rapid FRET Real-Time PCR Protocol for Simultaneous Quantitative Detection and Discrimination of Human Plasmodium Parasites. Bio-protocol 12(7): e4381. DOI: 10.21769/BioProtoc.4381.
分类
分子生物学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













