- EN - English
- CN - 中文
Phytophthora sojae Transformation Based on the CRISPR/Cas9 System
基于CRISPR/Cas9系统的大豆疫霉转化
发布: 2022年03月20日第12卷第6期 DOI: 10.21769/BioProtoc.4352 浏览次数: 3490
评审: Zhibing LaiFernando A Gonzales-ZubiateAnonymous reviewer(s)
Abstract
Phytophthora sojae is a model species for the study of plant pathogenic oomycetes. The initial research on gene function using Phytophthora was mainly based on gene silencing technology. Recently, the CRISPR/Cas9-mediated genome editing technology was successfully established in P. sojae and widely used in oomycetes. In this protocol, we describe the operating procedures for the use of CRISPR/Cas9-based genome editing technology and PEG-mediated stable transformation of P. sojae protoplasts. Two plasmids were co-transformed into P. sojae: pYF515 expressing Cas9 and the single guide RNA, and the homologous replacement vector of the candidate gene. Finally, the ORF of candidate gene were replaced with the ORF of the entire hygromycin B phosphotransferase gene (HPH), to achieve precise knockout.
Keywords: CRISPR/Cas9 (CRISPR/Cas9)Background
The probability of homologous recombination of Phytophthora itself is extremely low, and it is difficult to use methods based on homologous recombination to knock out genes. Therefore, a CRISPR/Cas9-based technology was developed to allow gene editing in P. sojae (Fang and Tyler, 2016). CRISPR/Cas9 is an adaptive immune response found in Bacteria and Archaea to prevent phage infection (Deveau et al., 2010; Garneau et al., 2010; Horvath and Barrangou, 2010). The CRISPR/Cas9 system consists of two components, namely Cas9 and single guide RNA (sgRNA) (Hsu et al., 2014). The single guide RNA (sgRNA) contains 20 nucleotides, which can mediate Cas9 to target the specific sequence region based on the principle of double-strand complementarity; Cas9 protein acts as a nuclease to cut specific DNA sites to produce double-strand break nicks (DSB).
In the presence of a homologous template (donor DNA), the cell uses the homologous template to repair, resulting in the homologous replacement of the target gene by the donor DNA to complete precise gene editing. Here, we provide detailed steps for the transformation of P. sojae based on the CRISPR/Cas9 system, including sgRNA design, Cas9-sgRNA plasmid construction, homologous replacement vector construction, P. sojae transformation, and detection of mutations, using Ps139767 as an example.
Materials and Reagents
Consumables
Sterile pipette tips (10 μL, 100 μL, 1,000 μL, 5 mL)
Petri dishes (60 mm × 15 mm, 90 mm ×15 mm)
250 mL Erlenmeyer flasks
Miracloth
Stainless steel laboratory spatulas
50 mL Falcon tubes
Microcentrifuge tubes (1.5 mL, 2.0 mL)
200 μL PCR tubes
20 mL Syringe
Beaker (50 mL, 200 mL)
Bacteria filter (0.45 nm)
Parafilm
Competent cells
E. coli JM109 competent cells (homemade)
Plasmids
pBluescript SK II+(pBS-SK II+)
pYF515 (constructed by Fang and Tyler, 2016) (Figure 1)
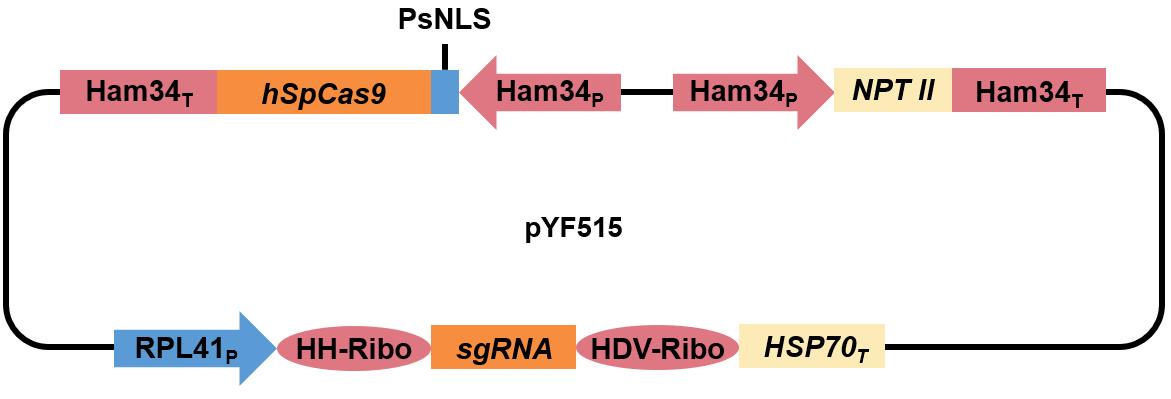
Figure 1. The pYF515 vector expresses both the Cas9 gene and the sgRNA. The expression of Cas9 and the selection marker NPT II is driven by the Ham34 promoter, and the transcription of sgRNA [including the flanking ribozyme, hammerhead ribozyme (HH ribozyme) and HDV ribozyme] is driven by the RPL41 promoter.
Primers (see Table 1)
Table 1. List of oligonucleotides (see Supplementary Information)
Primer name Sequence ( 5'→3' ) 139767-sgRNA1-F CTAGCTAGCCCGTCACTGATGAGTCCGTGAGGAC 139767-sgRNA1-R CTAGGGTCTCGAAACCAATGGTGACTACACCGTCAGACGAGCTTACTCGTTTCG 139767-sgRNA2-F CTAGCTAGCTAGTAGCTGATGAGTCCGTGAGGAC 139767-sgRNA2-R CTAGGGTCTCGAAACCCACTAGGTTGCAGTAGTAGGACGAGCTTACTCGTTTCG 139767-up1kb-F CTATAGGGCGAATTGGGTACCGCATTTCCGCAGTTCTCGTC 139767-up1kb-R GTCGCGGTGAGTTCAGGCATCGTCTACCTCCACTACGCG 139767-down1kb-F GTCCGAGGGCAAAGGAATAGACAAGCTACTCTTAGACTTTT 139767-down1kb-R AGGGAACAAAAGCTGGAGCTCAACGCAAGCACTGTCAAAGC HPH-F ATGCCTGAACTCACCGCGAC HPH-R CTATTCCTTTGCCCTCGGAC 139767-genome-F TTTGAAGACAAAAGCGGGCG 139767-genome-R TACTCGACGATACAGCACGC 139767-overup-F1 GGCCCGTGAATAAAACCCCT Hph-overup-R1 ACCATCGGCGCAGCTATTT Hph-overdown-F2 CGGGGATTCCCAATACGAGG 139767-overdown-R2 TAGCGTGTGTCAGATCCACC M13F GTAAAACGACGGCCAGT M13R CAGGAAACAGCTATGAC Strain
Phytophthora sojae strain P6497
E. coli JM109 (KangTi Life Technology, catalog number: KTSM107L)
Enzymes and buffers
BsaI and 10× Cutsmart Buffer (New England Biolabs, catalog number: R3733S)
NheI (New England Biolabs, catalog number: R3131S)
T4 DNA Ligase and 10× T4 DNA Ligase Buffer (Takara, catalog number: 2011A)
2× Phanta® Max Master Mix (Dye Plus) (Vazyme, catalog number: P525)
TaKaRa TaqTM,10× PCR Buffer(Mg2+ plus) and dNTP Mixture (Takara, catalog number: R001)
Kits
FastPure® Gel DNA Extraction Mini Kit (Vazyme, catalog number: DC301)
ClonExpress® ULtra One Step Cloning Kit (Vazyme, catalog number: C115)
FastPure® Plasmid Mini Kit (Vazyme, catalog number: DC201-01)
DNAsecure New Plant Genomic DNA Extraction Kit (TIANGEN BIOTECH, catalog number: DP320)
TaKaRa MiniBEST DNA Fragment Purification Kit Ver.4.0 (Takara, catalog number: 9761)
Antibiotics
Ampicillin (to a final concentration of 50 mg/mL) (Aladdin, catalog number: 69-52-3)
G418 (to a final concentration of 50 mg/mL) (Warbio, 108321-42-2)
Media (see Recipes)
Luria-Bertani agar plates + 50 mg/mL ampicillin
Luria-Bertani liquid medium
10% V8 agar plates +50 mg/mL G418
10% V8 liquid medium
Nutrient pea broth and agar medium (agar plates and liquid)
Mannitol-pea broth (agar plates and liquid)
Reagents
V8 vegetable juice (Campbell Soup Company, catalog number: 051000153159)
Frozen green peas
Lysing Enzymes (Sigma-Aldrich, catalog number: L1412)
CELLULYSIN Cellulase (Millipore, catalog number: 9012-54-8)
Bacto tryptone (OXOID, catalog number: 73049-73-7)
Yeast extract (Sigma-Aldrich, catalog number: 8013-01-2)
NaCl (Aladdin, catalog number: 7647-14-5)
Agar (Aladdin, catalog number: 9002-18-0)
CaCO3 (Sangon, catalog number: 471-34-1)
CaCl2 (Sangon, catalog number: 10043-52-4)
KCl (Sangon,catalog number: 7447-40-7)
PEG4000 (Aladdin, catalog number:25322-68-3)
Biotin (Aladdin, catalog number:58-85-5)
Folic acid (Aladdin, catalog number: 59-30-3)
L-inositol (Sigma-Aldrich, catalog number: 551-72-4)
Nicotinic acid (Aladdin, catalog number: 59-67-6)
Pyridoxine-HCl (Aladdin, catalog number: 58-56-0)
Riboflavin (Aladdin, catalog number: 83-88-5)
Thiamine-HCl (Aladdin, catalog number: 59-43-8)
FeC6H5O7·3H2O (Sangon, catalog number: 17217-76-4)
ZnSO4·7H2O (Sangon, catalog number: 7446-20-0)
CuSO4·5H2O (Sangon, catalog number: 7758-99-8)
MgSO4·H2O (Sangon, catalog number: 14567-64-7)
H3BO3 (Phytotech, catalog number: 10043-35-3)
MoO3 (Phytotech, catalog number: 1313-27-5)
KH2PO4 (Sangon, catalog number: 7778-77-0)
K2HPO4 (Sangon, catalog number: 7758-11-4)
KNO3 (Aladdin, catalog number: 7757-79-1)
D-sorbitol (Aladdin, catalog number: 50-70-4)
D-mannitol (Aladdin, catalog number: 69-65-8)
Glucose (Aladdin, catalog number: 50-99-7)
MgCl2·6H2O (Diamond, catalog number: 7791-18-6)
MES (Aladdin, catalog number: 4432-31-9)
Tris-HCl (Diamond, catalog number: 1185-53-1)
EDTA (Phytotech, catalog number: 60-00-4)
Hexadecyl trimethyl ammonium Bromide(CTAB) (Phytotech, catalog number: 57-09-0)
Solutions
0.8 M Mannitol solution, autoclave at 121°C for 20 min
0.5 M CaCl2 solution, sterilize using a 0.45-mm filter
0.5 M KCl solution, sterilize using a 0.45-mm filter
Luria-Bertani medium (see Recipes)
10% V8 agar (see Recipes)
Pea broth (see Recipes)
Nutrient pea broth and agar medium (see Recipes)
0.5 M MES-KOH solution (see Recipes)
W5 solution (see Recipes)
MMg solution (see Recipes)
Enzyme solution (see Recipes)
40% PEG solution (see Recipes)
Mannitol-pea broth (see Recipes)
2% CTAB solution (see Recipes)
Vitamin stock (see Recipes)
Trace elements (see Recipes)
Equipment
Pipettes (Eppendorf)
Rotating mixer (Select BioProducts, model: SBS550-2)
PCR machine (Bio-Rad, model: T100TM)
Light microscope (Leica, model: DM500)
Incubator (25°C) (Ningbo Jiangnan, model: HWS-1000)
Water bath (42°C) (SHELLAB, model: SWB7-2)
Tabletop centrifuge(4°C) (Thermofisher, model: ST8)
Incubator-shaker (37°C) (Crystal, model: IS-RSD3)
DNA electrophoresis apparatus (Bio-Rad, model: 1704469)
Software
sgRNA design software: EuPaGDT (http://grna.ctegd.uga.edu/)
Sequence alignment: SeqHunter2 (https://sourceforge.net/projects/seqhunter2)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Cao, J., Qiu, M., Ye, W. and Wang, Y. (2022). Phytophthora sojae Transformation Based on the CRISPR/Cas9 System. Bio-protocol 12(6): e4352. DOI: 10.21769/BioProtoc.4352.
分类
微生物学 > 微生物遗传学 > CRISPR-Cas9
分子生物学 > DNA > 转化
分子生物学 > DNA > 诱/突变
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













