- EN - English
- CN - 中文
RNA-mediated in vivo Directed Evolution in Yeast
RNA介导的酵母体内定向进化
发布: 2022年03月05日第12卷第5期 DOI: 10.21769/BioProtoc.4346 浏览次数: 3707
评审: Alessandro DidonnaAnonymous reviewer(s)
Abstract
Directed evolution is a powerful approach to obtain genetically-encoded sought-for traits. Compared to the prolonged adaptation regimes to mutations occurring under natural selection, directed evolution unlocks rapid screening and selection of mutants with improved traits from vast mutated sequence spaces. Many systems have been developed to search variant landscapes based on ex vivo or in vivo mutagenesis, to identify and select new-to-nature and optimized properties in biomolecules. Yet, the majority of such systems rely on tedious iterations of library preparation, propagation, and selection steps. Furthermore, among the relatively few in vivo directed evolution systems developed to mitigate handling of repetitive ex vivo steps, directed evolution of DNA is the standard approach. Here, we present the protocol for designing the transfer of genetic information from evolving RNA donors to DNA in baker’s yeast, using CRISPR- and RNA-assisted in vivo directed evolution (CRAIDE). We use mutant T7 RNA polymerase to introduce mutations in RNA donors, while incorporation into DNA is directed by CRISPR/Cas9. As such, CRAIDE offers an opportunity to study fundamental questions, such as RNA’s contribution to the evolution of DNA-based life on Earth.
Graphic abstract:
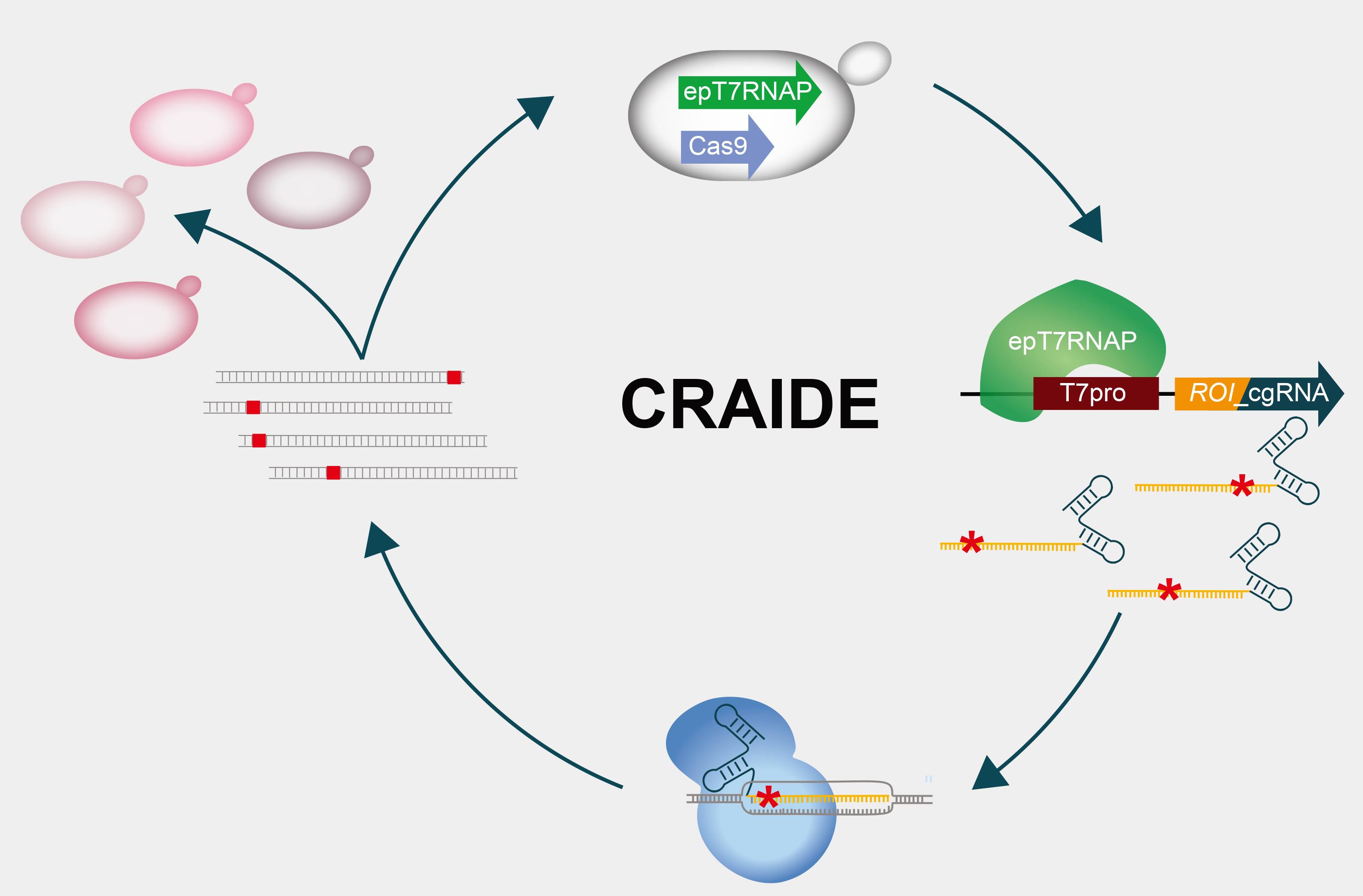
CRISPR- and RNA-assisted in vivo directed evolution (CRAIDE).
Background
The contribution of RNA to the evolution of DNA-based life is poorly understood. Detailed understanding of fundamental mechanisms and circumstances that facilitate information transfer from RNA to DNA are key to elucidate RNA’s role in evolution (Morris, 2015). Current technologies used in the laboratory to study evolution in vivo often focus on fixation of mutations directly into DNA by the use of error-prone and engineered polymerases (Ravikumar et al., 2014; Badran and Liu 2015; Moore et al., 2018; Halperin et al., 2018), and do not support studies on the possibility and frequency of evolving RNA molecules as templates for plasmid and genome editing. Here, we designed a synthetic genetically-encoded system to introduce evolving RNA donors into Cas9-targeted DNA loci in baker’s yeast, serving as a tool for investigating RNA-mediated evolution on plasmids and genomic contexts. This system called CRISPR- and RNA-assisted in vivo directed evolution (CRAIDE) builds upon previous reports, showing that transcripts delivered in vivo can support RNA-DNA repair of induced DNA double-strand breaks (DSBs) in yeast RNase H knock-out strains (Keskin et al., 2014 and 2016). Extending from these seminal findings, CRAIDE makes use of an orthogonal error-prone T7 RNA polymerase to control the expression of chimeric guide RNAs (cgRNAs), which encode both a seed sequence and an evolved template for repair at user-defined loci targeted by Cas9 (Jensen et al., 2021). We envision CRAIDE to be useful for i) studies on the basics of RNA-templated DNA repair during evolution, ii) the development and further optimization of design principles for targeted and efficient genome engineering, as well as iii) in vivo directed evolution by use of a simple set of plasmid-based designs.
Materials and Reagents
1.5 mL safe-lock tubes (Sigma-Aldrich, catalog number: 0030 120.086)
Petri dishes (TH. Geyer, Labsolute, catalog number: 7696405)
14 mL culture tubes (Fisher Scientific, Greiner Bio-One, catalog number: 07-000-212)
PCR tubes (Sarstedt Inc., 0.2 mL PCR tube multiply μstrip 8 neutral PCR perf, catalog number: 72.985.002)
Inoculation loops (Deltalab S.L., 10 μL, catalog number: 302704)
Cuvettes (BrandTech Scientific Inc., catalog number: 7590 15)
Whatman paper (Sigma-Aldrich, catalog number: WHA10300045)
Replication block (Sigma-Aldrich, catalog number: Z363391-1EA)
50 mL syringe (VWR, catalog number: 89215-240)
0.2 μm single use filter unit (TH. Greyer, Labsolute, catalog number: 7699822)
Yeast (CEN.PK2-1C, EUROSCARF) (see Table 1)
Lithium acetate (Sigma-Aldrich, catalog number: 517992)
PEG 3350 (Sigma-Aldrich, catalog number: 1546547)
ssDNA (Sigma-Aldrich, single stranded deoxyribonucleic acid from salmon testes, catalog number: D9156)
Sodium acetate, 3 M, pH 5.2 (Merck Millipore, catalog number: 567422)
100% ethanol (VWR, catalog number: 20821.310)
70% ethanol (VWR, catalog number: 83801.360)
NucleoSpin gel and PCR clean-up kit (Macherey-Nagel, catalog number: 740609.50)
Yeast synthetic drop-out medium supplements without leucine, tryptophan, and uracil (Sigma-Aldrich, catalog number: Y1771)
Yeast synthetic drop-out medium supplements without histidine (Sigma-Aldrich, catalog number: Y1751)
Yeast synthetic drop-out medium supplements without leucine (Sigma-Aldrich, catalog number: Y1376)
YPD Broth (ThermoFisher Scientific, catalog number: A1374501)
D-(+)-Galactose (Sigma-Aldrich, catalog number: G0625)
D-(+)-Glucose (Sigma-Aldrich, catalog number: G8270)
Yeast nitrogen base without amino acids (Sigma-Aldrich, catalog number: Y0626)
Yeast nitrogen base without amino acids and ammonium sulfate (Sigma-Aldrich, catalog number: Y1251)
L-Glutamic acid monosodium salt monohydrate (Sigma-Aldrich, catalog number: 49621)
Agar (Merck Millipore, catalog number: 05040)
Nourseothricin (Jena Bioscience, catalog number: AB-102XL)
GeneRuler 1 kb DNA ladder (Thermo Scientific, catalog number: SM0311)
Agarose (Sigma-Aldrich, catalog number: A4718)
RedSafe (iNtRON Biotechnology, catalog number: 21141)
DNA gel loading dye 6× (Thermo Scientific, catalog number: R0611)
OneTaq 2× Master Mix with standard buffer (New England BioLabs, catalog number: M0482S)
Oligonucleotides (IDT) (see Table 3)
Plasmids (see Table 2)
FastDigest NotI (ThermoFisher Scientific, catalog number: FD0593)
FastDigest buffer (ThermoFisher Scientific, catalog number: B64)
TAE Buffer (Tris-Acetate-EDTA) 50× (ThermoFisher Scientific, catalog number: B49)
200 mg/mL nourseothricin solution (see Recipes)
Synthetic complete drop-out media (see Recipes)
20% (w/v) glucose and galactose solutions (see Recipes)
Table 1. Yeast strains.
Strain Genotype Plasmids Parental strain Reference CEN.PK2-1C MATa ura3-52 his3∆1 leu2-3_112 trp1-289 MAL2-8c SUC2 N/A N/A EUROSCARF Sc35 MATa ura3-52 his3∆1 leu2-3_112 trp1-289 MAL2-8c SUC2 pEDJ391 CEN.PK2-1C Jensen et al., 2021 Sc36 MATa ura3-52 his3∆1 leu2-3_112 trp1-289 MAL2-8c SUC2 rnh1∆ rnh201∆ N/A CEN.PK2-1C Jensen et al., 2021 Sc43 MATa ura3-52 leu2-3_112 trp1-289 MAL2-8c SUC2 rnh1∆ rnh201∆ his3∆ pEDJ391 Sc40 Jensen et al., 2021 Sc138 MATa ura3-52 leu2-3_112 trp1-289 MAL2-8c SUC2 rnh1∆ rnh201∆ his3∆ HIS3_23∆29-XII-5 N/A Sc43 Jensen et al., 2021 Sc146 MATa ura3-52 leu2-3_112 trp1-289 MAL2-8c SUC2 rnh1∆ rnh201∆ his3∆ HIS3_23∆29-XII-5 pEDJ333; pMLB10; pEDJ508 Sc138 Jensen et al., 2021 Sc147 MATa ura3-52 leu2-3_112 trp1-289 MAL2-8c SUC2 rnh1∆ rnh201∆ his3∆ HIS3_23∆29-XII-5 pEDJ333; pMLB10; pEDJ400 Sc138 Jensen et al., 2021 Table 2. Plasmids.
Plasmid Contents Genetic marker Antibiotic marker Origin of replication Addgene ID pEDJ8 SNR52p:sgRNA_RNH1∆:SUP4t_SNR52:sgRNA_RNH201∆:SUP4t URA3 Ampicillin 2μ 177271 pEDJ332 SNR52p:sgRNA_HIS3-KO:SUP4t Nourseothricin resistance Ampicillin 2μ N/A pEDJ333 GAL1p:Cas9:CYC1t LEU2 Ampicillin Cen/ARS 177272 pEDJ391 TEF1p:Cas9:CYC1t LEU2 Ampicillin Cen/ARS N/A pEDJ400 USER cloning site URA3 Ampicillin 2μ 177269 pEDJ437 USER cloning site HIS3 Ampicillin 2μ 177270 pEDJ506 ADH1t:HIS3_23∆29-XII-5:<-PGK1p N/A (no marker) Ampicillin 2μ 177274 pEDJ508 ADH1t:T7p:cgRNA_HIS3_stop:tZ URA3 Ampicillin 2μ 177275 pMLB10 GAL1p:T7RNAP(F11L;T613A):CYC1t TRP1 Ampicillin 2μ 177273 pCfB3050 SNR52p:sgRNA_XII-5:SUP4t Nourseothricin resistance Ampicillin 2μ 73292 Table 3. Oligos.
Oligo Sequence F_RNH1_KO CCTACTGTATTGTACTTGAAACAGG R_RNH1_KO GCCACTCCGAAGGTTTGAAAGTCG F_RNH201_KO GTACCCCCCACGGTAGAAGCATC R_RNH201_KO CAATTATCTAGGGTTCTCAGC F_HIS3_KO ACTCTTGGCCTCCTCTAGTACAC R_HIS3_KO CATGTATATATATCGTATGCTGCAG F_Seq CAAGTTCTTAGATGCTTTC R_Seq CTACATAAGAACACCTTTGGTG RNH1_90-mer AAAGTGTCACTCCTTGCTTATCGAAGGAACTATCGATTCCTAATTTGCTGTTTTTTGCTTGGCTTCTTACTGGACCTGTTGTACCTGTAA RNH201_90-mer AAAAACCTTGAAAACAACTACTGCACACCAAATTGATACGATTAATCACCTCCAGTTTGCACATACTATATATCTTCATGTAATACTACA HIS3_90-mer AAGAATATACTAAAAAATGAGCAGGCAAGATAAACGAAGGCAAAGTGACACCGATTATTTAAAGCTGCAGCATACGATATATATACATGT
Equipment
-80°C freezer (Thermo Scientific, Forma 88000 Series, catalog number: 88-500A)
30°C shaking incubator (Thermo Scientific, MaxQ 8000, model: SHKE8000)
Stationary incubator (Thermo Scientific, Heratherm, catalog number: 51028112)
Safety cabinet (Thermo Scientific, Safe 2020, catalog number: 51026637)
Autoclave (CertoClav, CertoClav Multicontrol, catalog number: 8502312A)
Centrifuge (Thermo Scientific, Multifuge X1 Centrifuge, catalog number: 75004211)
Benchtop centrifuge (VWR, Micro Star 17, catalog number: 521-1646)
Refrigerated centrifuge (VWR, Micro Star 17R, catalog number: 521-1647)
Spectrophotometer (IMPLEN, model: NanoPhotometer P300)
Thermomixer (VWR, model thermomixer comfort)
Vortexer (Scientific Industries, model: Vortex-Genie 2)
Water purification system (VWR, PURELAB flex 2, catalog number: 89204-092)
Molecular Imager Gel Doc (Bio-Rad Laboratories, catalog number: 1708195)
DNA electrophoresis chamber (Bio-Rad Laboratories, catalog number: 1704487EDU)
Fixed-Height Comb (Bio-Rad Laboratories, catalog number: 1704465EDU)
Sub-Cell GT UV-Transparent Mini-Gel tray (Bio-Rad Laboratories, catalog number: 1704435EDU)
Mini-Gel caster (Bio-Rad Laboratories, catalog number: 1704422EDU)
PowerPac basic power supply (Bio-Rad Laboratories, catalog number: 1645050)
Thermal cycler (Bio-Rad Laboratories, S1000, catalog number: 1852148)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Jensen, E. D. and Jensen, M. K. (2022). RNA-mediated in vivo Directed Evolution in Yeast. Bio-protocol 12(5): e4346. DOI: 10.21769/BioProtoc.4346.
分类
微生物学 > 微生物遗传学 > CRISPR-Cas9
细胞生物学 > 细胞工程 > CRISPR-cas9
分子生物学 > DNA > DNA 损伤和修复
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












