- EN - English
- CN - 中文
Laser Microirradiation and Real-time Recruitment Assays Using an Engineered Biosensor
利用工程生物传感器的激光微照射和实时招募分析
发布: 2022年03月05日第12卷第5期 DOI: 10.21769/BioProtoc.4337 浏览次数: 3783
评审: Chiara AmbrogioAnonymous reviewer(s)
Abstract
Double-strand breaks (DSBs) are lesions in DNA that, if not properly repaired, can cause genomic instability, oncogenesis, and cell death. Multiple chromatin posttranslational modifications (PTMs) play a role in the DNA damage response to DSBs. Among these, RNF168-mediated ubiquitination of lysines 13 or 15 at the N-terminal tail of histone H2A (H2AK13/15Ub) is essential for the recruitment of effectors of both the non-homologous end joining (NHEJ) and the homologous recombination (HR) repair pathways. Thus, tools and techniques to track the spatiotemporal dynamics of H2AK13/15 ubiquitination at DNA DSBs are important to facilitate studies of DNA repair. Previous work from other groups used the minimal focus-forming region (FFR) of the NHEJ effector 53BP1 to detect H2AK15Ub generated upon damage induced by gamma or laser irradiation in live cells. However, 53BP1-FFR only binds nucleosomes modified with both H2AK15Ub and dimethylation of lysine 20 on histone H4 (H4K20me2); thus, 53BP1-FFR does not recognize H2AK13Ub–nucleosomes or nucleosomes that contain H2AK15Ub but lack methylation of H4K20 (H4K20me0). To overcome this limitation, we developed an avidity-based sensor that binds H2AK13/15Ub without dependence on the methylation status of histone H4K20. This sensor, called Reader1.0, detects DNA damage-associated H2AK13/15Ub in live cells with high sensitivity and selectivity. Here, we present a protocol to detect the formation of H2AK13/15Ub at laser-induced DSBs using Reader1.0 as a live-cell reporter for this histone PTM.
Graphic abstract:
Background
DNA double-strand breaks (DSBs) are a threat to genome integrity and stability. DSBs can be generated in response to external agents (e.g., irradiation, radiomimetic chemicals) or formed as byproducts of normal cell metabolism (e.g., oxidative stress, DNA replication errors). To deal with DSBs, eukaryotic cells use complex repair machinery equipped with protein effectors responsible for detecting, signaling, and promoting the repair of these lesions. Defects in DNA DSB repair can lead to mutations or chromosomal aberrations and, ultimately, contribute to the onset of cancer and neurodegenerative diseases (Jackson and Bartek, 2009; Ciccia and Elledge, 2010; Polo and Jackson, 2011).
In response to DNA DSBs, the serine/threonine kinase ataxia-telangiectasia mutated (ATM) is activated and promotes the phosphorylation of various proteins in chromatin surrounding the damage sites. These phosphorylation substrates include histone variant H2AX in vertebrates. Phosphorylated H2AX (γH2AX) leads to the sequential recruitment of mediator of DNA damage checkpoint protein 1 (MDC1) and the E3 ubiquitin ligases RNF8 and RNF168. RNF168 specifically ubiquitinates lysines 13 or 15 at the N-terminal tail of histone H2A (H2AK13/15Ub) (Mattiroli et al., 2012), which in turn can recruit effectors of non-homologous end joining (NHEJ) or homologous recombination (HR)-mediated repair (Mattiroli and Penengo, 2021). The NHEJ repair factor 53BP1 is recruited by nucleosomes containing both H4K20me2 and H2AK15Ub (Fradet-Turcotte et al., 2013; Wilson et al., 2016; Hu et al., 2017). In contrast, the BRCA1/BARD1 complex promotes the HR pathway and is recruited by nucleosomes modified with H2AK13Ub or H2AK15Ub, only if H4K20 is not methylated (H4K20Me0) (Becker et al., 2021; Hu et al., 2021). The minimal focus-forming region (FFR) of 53BP1 has been previously shown to localize to DSBs induced by gamma or laser irradiation, and thereby report local increases in H2AK15Ub at DNA lesions (Fradet-Turcotte et al., 2013; dos Santos Passos et al., 2021). However, structural and biochemical studies have demonstrated that 53BP1 binding to the nucleosomes requires both H2AK15Ub and H4K20Me2 modifications. The H4K20Me2 modifications are enriched in the G1 and early S-phases of the cell cycle (Fradet-Turcotte et al., 2013; Wilson et al., 2016; Hu et al., 2017; Michelena et al., 2021).
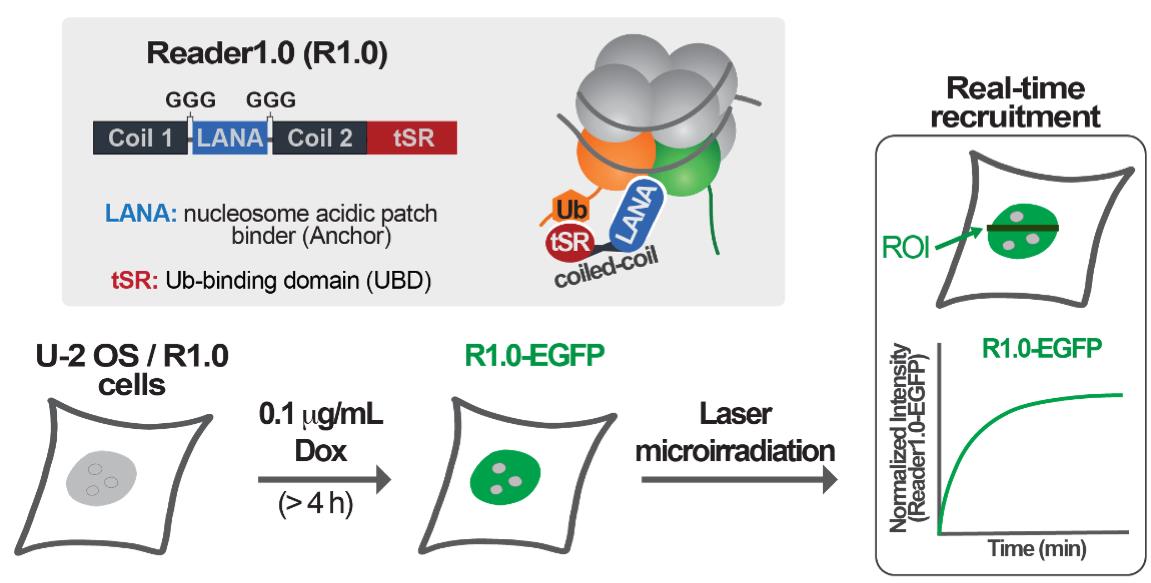
In recent work, we developed and validated an avidity-based sensor to detect H2AK13/15Ub-nucleosomes. This sensor, which we call Reader1.0, can bind nucleosomes containing either H2AK13Ub or H2AK15Ub in vitro, and it localizes to sites of RNF168-mediated ubiquitination in mammalian cells. In addition, Reader1.0 binding to H2AK13/15Ub-nucleosomes does not depend on the methylation status of histone H4K20; i.e., Reader1.0 recognizes H2AK13/15Ub-nucleosomes containing either H4K20me0 or H4K20me2. When fused to fluorescent proteins and expressed at tightly-controlled levels, Reader1.0 can be used to monitor the dynamics of H2AK13/15Ub in live cells over long time intervals without causing significant perturbation of cell growth rate or DNA repair kinetics (dos Santos Passos et al., 2021).
The protocol presented here describes how to measure the recruitment of readers of RNF168-dependent H2AK13/15 ubiquitination to DNA DSBs. To rapidly generate local high concentrations of H2AK13/15Ub, laser microirradiation was used to induce DNA damage in the nuclei of U-2 OS cells. The main advantages of this method are the abilities to target defined nuclear volumes and to generate similar amounts of DNA damage in different nuclei within a cell population (Bekker-Jensen et al., 2006; Lukas et al., 2003). By co-expressing mCherry-53BP1-FFR and Reader1.0-EGFP, we demonstrated that our designed sensor Reader1.0 and 53BP1-FFR both redistribute to laser-induced DNA lesions with similar kinetics. In sum, this protocol describes the detection of H2AK13/15Ub at DNA DSBs in live cells by fluorescence confocal microscopy.
Materials and Reagents
60 mm tissue culture dishes (Genesse Scientific, catalog number: 26-260)
35-mm glass-bottom dishes (MatTek, catalog number: P35G-1.5-14-C)
Phosphate buffered saline (PBS; Corning, catalog number: 21-040-CV)
TrypLETM express enzyme (Gibco, catalog number: 12604021)
Dulbecco's modified Eagle's medium (DMEM; Corning, catalog number: 15-013-CV)
FluoroBriteTM DMEM (Gibco, catalog number: A1896701)
Opti-MEMTM reduced serum medium (Gibco, catalog number: 11058021)
Penicillin-Streptomycin 100× solution (10,000 U/mL; HyClone, catalog number: SV30010)
L-Glutamine 200 mM solution (HyClone, catalog number: SH30034.01)
Fetal bovine serum (FBS; Sigma-Aldrich, catalog number: F0926)
LipofectamineTM 3000 transfection reagent (ThermoFisher, catalog number: L3000015)
Dimethyl sulfoxide (DMSO, Fisher Scientific, catalog number: BP231-100)
Doxycycline (Dox) hydrochloride (Fisher Scientific, catalog number: BP2653-1)
U-2 OS cells stably expressing Reader1.0-EGFP under the control of a Dox-inducible promoter (dos Santos Passos et al., 2021)
Plasmid mCherry-BP1-2 pLPC-Puro (Addgene, plasmid number: 12259)
Growth medium (see Recipes)
Live-cell medium (see Recipes)
Equipment
CO2 incubator (Thermo Fisher Scientific, model: Heracell VIOS 160i)
Bead bath 37°C incubator (Lab Armor, model: 74300-706)
Vortex mixer (Fisher Scientific, model: Genie 2, catalog number: 12-812)
Confocal laser scanning microscope (Carl-Zeiss, model: LSM 880) operated with Zen Black 2.3 software (v.14.0.9.201) and equipped with a Plan-Apochromat 63×/1.40 oil-immersion objective
Stage Incubator for Live-Cell Imaging (PECON, PM 2000 RBT)
Software
Zen Black 2.3 software (v.14.0.9.201; Carl-Zeiss)
Zen Blue 2.3 software (v.2.3.69.1000; Carl-Zeiss)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- dos Santos Passos, C., Cohen, R. E. and Yao, T. (2022). Laser Microirradiation and Real-time Recruitment Assays Using an Engineered Biosensor. Bio-protocol 12(5): e4337. DOI: 10.21769/BioProtoc.4337.
- dos Santos Passos, C., Choi, Y.-S., Snow, C. D., Yao, T., Cohen, R. E. (2021). Design of genetically encoded sensors to detect nucleosome ubiquitination in live cells. J Cell Biol 220e201911130.
分类
生物工程 > 合成生物学
分子生物学 > DNA > DNA 损伤和修复
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link