- EN - English
- CN - 中文
Activity-based Crosslinking to Identify Substrates of Thioredoxin-domain Proteins in Malaria Parasites
基于活性的交联识别疟原虫中硫氧还蛋白结构域蛋白的底物
发布: 2022年02月20日第12卷第4期 DOI: 10.21769/BioProtoc.4322 浏览次数: 2891
评审: Alexandros AlexandratosMigla MiskinyteDarrell CockburnPetru-Iulian Trasnea
Abstract
Malaria remains a major public health issue, infecting nearly 220 million people every year. The spread of drug-resistant strains of Plasmodium falciparum around the world threatens the progress made against this disease. Therefore, identifying druggable and essential pathways in P. falciparum parasites remains a major area of research. One poorly understood area of parasite biology is the formation of disulfide bonds, which is an essential requirement for the folding of numerous proteins. Specialized chaperones with thioredoxin (Trx) domains catalyze the redox functions necessary for breaking incorrect and forming correct disulfide bonds in proteins. Defining the substrates of these redox chaperones is difficult and immunoprecipitation based assays cannot distinguish between substrates and interacting partners. Further, the substrate or client interactions with the redox chaperones are usually transient in nature. Activity based crosslinkers that rely on the nucleophilic cysteines on Trx domains and the disulfide bond forming cysteines on clients provide an easily scalable method to trap and identify the substrates of Trx-domain containing chaperones. The cell permeable crosslinker divinyl sulfone (DVSF) is active only in the presence of nucleophilic cysteines in proteins and, therefore, traps Trx domains with their substrates, as they form mixed disulfide bonds during the course of their catalytic activity. This allows the identification of substrates that rely on Trx activity for their folding, as well as discovering small molecules that interfere with Trx domain activity.
Graphic abstract:
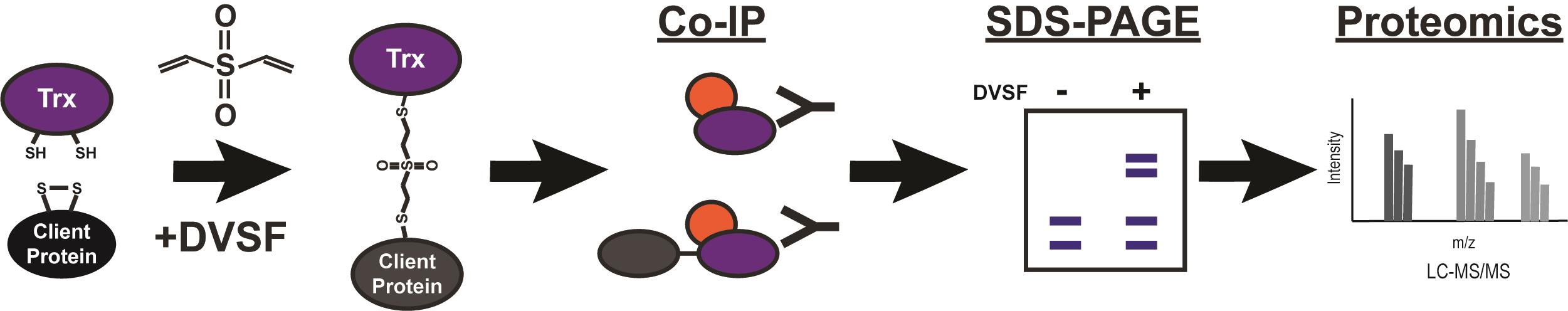
Identification of thioredoxin domain substrates via divinylsulfone crosslinking and immunoprecipitation-mass spectrometry.
Background
Disulfide bonds are crucial for the activity and regulation of many proteins; therefore, disulfide bond formation and reduction is a critical aspect of cellular life. Members of the thioredoxin (Trx) superfamily use the nucleophilic cysteines of their “CXXC” active sites to facilitate the formation and reduction of disulfide bonds (Lu and Holmgren, 2014). Given their importance for cell survival, thioredoxin-domain proteins and the thioredoxin system have been proposed as drug targets against apicomplexan parasites (Becker et al., 2000; Krnajski et al., 2001; Andricopulo et al., 2006; Kehr et al., 2010; Biddau et al., 2018; Biddau and Sheiner, 2019; Cobb et al., 2021). Despite being essential and their potential as drug targets, the substrates of these Trx-domain proteins are often ill-defined, as it can be difficult to discern specific substrates from other interacting partners.
One strategy for identifying the substrates of Trx domains relies on the active site mode of action: the first cysteine of the “CXXC'' active site forms a mixed disulfide bond with a client protein, and the sulhydryl group of the second cysteine is used to resolve that bond. Mutagenesis of the second cysteine, often to alanine or serine, prevents the reduction of the bond between the Trx-domain and client protein, trapping the two proteins together (Jessop et al., 2007; Oka et al., 2013). Subsequently, mass spectrometry can be used to identify the substrate. A major disadvantage to this approach is that it requires modification to the active sites of endogenous enzymes, that may be essential, or exogenous expression of the mutated enzymes, both of which may prove lethal for the organism. Particularly in organisms like Plasmodium falciparum, which is haploid for the majority of its lifecycle and carries single copies of genes for essential Trx-domain proteins, this approach may prove especially difficult.
Given the challenges inherent to a genetic approach, activity based chemical crosslinking of Trx-domains to their substrates is a straightforward and powerful option for identifying those substrates. Divinyl sulfone (DVSF) is a bifunctional, electrophilic, and cell permeable crosslinker shown to have remarkable specificity for nucleophilic cysteines, like those within Trx-domain active sites (West et al., 2011). Treatment of cells with DVSF results in irreversible crosslinking between redox-active cysteines, and it has been used to trap and identify substrates of Trx-domain proteins and other redox-active enzymes in yeast and human cells (Naticchia et al., 2013; Allan et al., 2016; Araki et al., 2017). Demonstrating the power of this activity-based technique for identifying Trx-domain substrates in a non-model organism, we used DVSF to trap and identify substrates of Trx-domain proteins in the endoplasmic reticulum (ER) of P. falciparum parasites (Cobb et al., 2021). We also demonstrated its specificity for redox-active nucleophilic cysteines, as other cysteine-containing proteins did not react with DVSF, and further validated our results by demonstrating ablation of crosslinking, by mutation of reactive cysteine residues to alanine residues (Cobb et al., 2021). In this protocol, we describe the use of DVSF to trap substrates of ER-localized Trx-domain proteins in P. falciparum and the identification of these trapped substrates via mass spectroscopy. Given the robust nature of this protocol, it will be of use for similar experiments in diverse organisms and within multiple cellular compartments. Because this approach can be utilized with minimal genetic modification, requiring only the addition of an affinity tag to the Trx-domain protein if a suitable antibody is not otherwise available, this protocol will allow others working with difficult organisms to study the interactions between Trx-domains and their substrates.
Materials and Reagents
15 mL centrifuge tubes (Genesee Scientific, catalog number: 28-103)
1.7 mL microtubes (Genesse Scientific, catalog number: 28-281)
Filter tip pipette tips: 10 μL (Fisher Scientific, catalog number: 02-707-473), 200 μL (Fisher Scientific, catalog number: 02-707-478), 1 mL (Fisher Scientific, catalog number: 2707480)
Serological pipettes: 5 mL (Genesee Scientific, catalog number: 12-102), 10 mL Genesee Scientific, catalog number: 12-104)
TPP Tissue culture plate, 100 mm (MidSci, catalog number: TP92406)
O+ AS-1 Packed Cells (Interstate Blood Bank, Memphis, TN)
IRDye® 800CW anti-mouse (Li-Cor, catalog number: 926-32210 used at 1:20,000 for Western blot)
IRDye® 680RD anti-rabbit (Li-Cor, catalog number: 926-68071, used at 1:20,000 for Western blot)
Rabbit anti-HA antibody (Life Technologies, catalog number: 715500, used at 1:100 for Western blot)
Mouse anti-PMV antibody (Gift from Daniel Goldberg, Washington University School of Medicine, used at 1:400 for Western blot)
V5-Tag (D3H8Q) Rabbit mAb (Cell Signaling Tech, Inc., catalog number: 13202S, used at 1:1,000 for Western blot)
Anti-HA Magnetic Beads (Thermo Fisher Scientific/PierceTM, catalog number: 88836)
Divinyl sulfone, 97% stab. with 0.05% hydroquinone (Thermo Fisher Scientific/Alfa Aesar, catalog number: L12827)
Saponin Quillaja sp., Sapogenin content 20-35% (Sigma, catalog number: S4521-10G)
Protein sample loading buffer, 4× (Li-Cor, catalog number: 928-40004)
Chameleon Duo pre-stained protein ladder (Li-Cor, catalog number:928-60000)
2-mercaptoethanol (Fisher Scientific, catalog number: 21985023)
10× Tris/Glycine/SDS buffer (Bio-Rad, catalog number: 161-0732)
4-20% Mini-PROTEAN® TGXTM precast protein gels (Bio-Rad, catalog number: 456-1093)
InterceptTM blocking buffer (Li-Cor, catalog number: 927-70003)
Odyssey® nitrocellulose membrane (Li-Cor, catalog number: 926-31092)
Halt protease inhibitor cocktail (Thermo Fisher Scientific/Life Technologies, catalog number: 78429)
HA synthetic peptide (Thermo Fisher Scientific/Life Technologies, catalog number: 26184)
Simply Blue SafeStain (Thermo Fisher Scientific/Life Technologies, catalog number: LC6060)
Tris-HCl (Fisher Bioreagents, catalog number: 1185-53-1)
KCl (Sigma, catalog number: 7447-40-7)
EDTA (Fisher Bioreagents, catalog number: 60-00-4)
NP-40 (Sigma, catalog number: 9002-93-1)
Na2HPO4 (Millipore Sigma, catalog number: 1065861000)
KH2PO4 (Millipore Sigma, catalog number: 529568)
NaCl (Fisher Scientific, catalog number: BP358212)
TRIS base (Fisher Scientific, catalog number: YBP152500)
Glycine (Fisher Scientific, catalog number: ALF-036435-A1)
Methanol (Fisher Scientific, catalog number: A4081)
Tween-20 (Fisher Scientific, catalog number: BP337-100)
Immunoprecipitation extraction buffer (see Recipes)
Immunoprecipitation binding buffer (see Recipes)
Phosphate buffered saline (PBS), pH 7.4 (see Recipes)
Protein loading dye (see Recipes)
10× Transfer buffer (see Recipes)
1× Transfer buffer (see Recipes)
0.1% Tween-20, in 1× PBS (see Recipes)
10× Tris-buffered saline (TBS) (see Recipes)
2 mg/mL HA peptide (see Recipes)
Equipment
SorvallTM LegendTM XTR Centrifuge (Thermo Fisher Scientific, model: 75004506)
Odyssey® CLx Imaging System (Li-Cor, model: 9140)
DynaMagTM-2 Magnet (Life Technologies, model: 12321D)
Branson S450D Digital Sonifier (Thermo Fisher Scientific, model: 101-063-593)
Mini-PROTEAN Tetra Cell System (Bio-Rad, catalog number: 1658005EDU)
PowerPacTM Basic Power Supply (Bio-Rad, catalog number: 1645050)
FormaTM Series II Water-Jacketed CO2 incubator (ThermoFisher Scientific, catalog number: 3110)
Software
Image Studio 5.x CLx (Li-Cor, https://www.licor.com/bio/image-studio-lite/)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Cobb, D. W., Woods, G. S. and Muralidharan, V. (2022). Activity-based Crosslinking to Identify Substrates of Thioredoxin-domain Proteins in Malaria Parasites. Bio-protocol 12(4): e4322. DOI: 10.21769/BioProtoc.4322.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 活性
生物化学 > 蛋白质 > 相互作用 > 交联
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link