- EN - English
- CN - 中文
Amber Suppression Technology for Mapping Site-specific Viral-host Protein Interactions in Mammalian Cells
哺乳动物细胞中定位特异性病毒-宿主蛋白相互作用的琥珀抑制技术
发布: 2022年02月05日第12卷第3期 DOI: 10.21769/BioProtoc.4315 浏览次数: 4219
评审: Khyati Hitesh ShahHeng SunWenn-Chyau Lee
Abstract
Probing the molecular interactions of viral-host protein complexes to understand pathogenicity is essential in modern virology to help the development of antiviral therapies. Common binding assays, such as co-immunoprecipitation or pull-downs, are helpful in investigating intricate viral-host proteins interactions. However, such assays may miss low-affinity and favour non-specific interactions. We have recently incorporated photoreactive amino acids at defined residues of a viral protein in vivo, by introducing amber stop codons (TAG) and using a suppressor tRNA. This is followed by UV-crosslinking, to identify interacting host proteins in live mammalian cells. The affinity-purified photo-crosslinked viral-host protein complexes are further characterized by mass spectrometry following extremely stringent washes. This combinatorial site-specific incorporation of a photoreactive amino acid and affinity purification-mass spectrometry strategy allows the definition of viral-host protein contacts at single residue resolution and greatly reduces non-specific interactors, to facilitate characterization of viral-host protein interactions.
Graphic abstract:
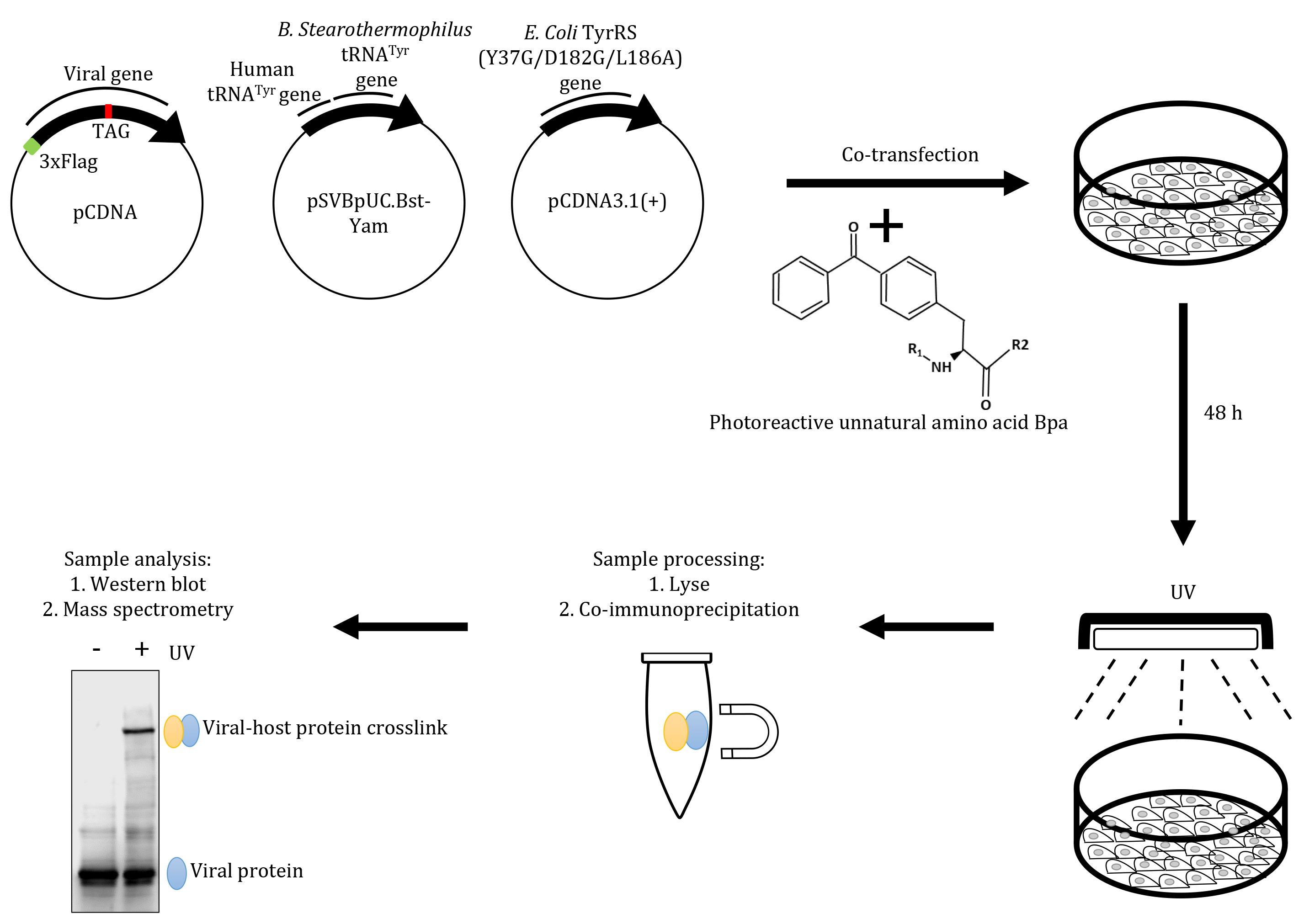
Schematic overview of the virus-host interaction assay based on an amber suppression approach. Mammalian cells grown in Bpa-supplemented medium are co-transfected with plasmids encoding viral sequences carrying a Flag tag, a (TAG) stop codon at the desired position, and an amber suppressor tRNA (tRNACUA)/aminoacyl tRNA synthetase (aaRS) orthogonal pair. Cells are then exposed to UV, to generate protein-protein crosslinks, followed by immunoprecipitation with anti-Flag magnetic beads. The affinity-purified crosslinks are probed by western blot using an anti-Flag antibody and the crosslinked host proteins are characterised by mass spectrometry.
Background
In situ binding assays such as pull-downs or affinity purification coupled with mass spectrometry are reliable approaches to study viral-host protein interactions. However, it is important to recognize the limitations of these methods, including lack of specificity, which can lead to false-positives in the interactome. To minimize non-specific binding artifacts, we have designed an interaction assay based on ectopic expression of an engineered viral protein in mammalian cells, in the presence of a suppressor tRNA (tRNASup)/aminoacyl tRNA synthetase (aaRS) orthogonal pair and growth medium supplemented with an unnatural photoreactive amino acid (Ye et al., 2008; Neumann-Staubitz and Neumann, 2016; Xiao and Schultz, 2016; Chin, 2017). The amber codon TAG is first introduced at the desired residues within the viral protein. During ectopic expression of these mutated viral proteins in the presence of the tRNASup/aaRS pair in mammalian culture, the aaRS charges an unnatural photocrosslinkable amino acid to the tRNASup, which is then incorporated at the position of the amber codon. The cells expressing the viral protein containing the unnatural amino acid are then exposed to UV light, which induces covalent photocrosslinks to host proteins present in close proximity to the viral protein. The crosslinked complex is stable and can be affinity-purified in stringent conditions, to eliminate non-specific interactions and be characterised by mass spectrometry. Over the years, the approach has been applied to interactions between cellular proteins (Hino et al., 2005; Kobbi et al., 2016; Wu et al., 2020). We have adapted this approach to interrogate the interaction between host-cell proteins with the viral protein HSV-1 ICP22 (Isa et al., 2021), and this is applicable to other viral proteins.
Materials and Reagents
1.5 mL microcentrifuge tube (Starlab, catalog number: E1415-1510)
Pipette tip (10 μL, 200 μL, and 1,000 μL) (Sarstedt, catalog numbers: 70.3010, 70.760.3010, 70.3050.200)
Cell culture Petri dishes (Starlab, catalog number: CC7682-3394)
Mammalian expression vector carrying N-terminally 3× FLAG-tagged proteins, as for example p3× FLAG-CMVTM-8 (Merck, catalog number: E9908)
pSVBpUC.Bst-Yam suppressor tRNA plasmid, a generous gift from Dr. Shixin Ye-Lehman (read as tRNASup). Gene cloning was described by Ye et al. (2008)
pCDNA3.1(+).aminoacyl-tRNA synthetase specific for Bpa, a generous gift from Dr. Shixin Ye-Lehman (read as aaRS). Gene cloning was described by Ye et al. (2008)
HEK293 cells (ATCC®, catalog number: CRL-1573TM)
Anti-Flag M2 Magnetic beads (Sigma-Aldrich, catalog number: M8823; -20°C)
Product H-L-Bpa-OH (p-benzoyl-L-phenylalanine) (IRIS Biotech, catalog number: HAA6010) (read as Bpa)
QuikChange multi site-directed mutagenesis kit (Agilent Technologies, catalog number: 200514)
DMEM (Sigma-Aldrich, catalog number: D6429; 4°C)
jetPRIME® transfection reagent (PolyPlus, catalog number: 114-15; 4°C)
cOmpleteTM EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich, catalog number: 5056489001, 4°C). Store dissolved stock solution at -20°C
Benzonase (Merck Chemicals, catalog number: 70664-3; -20°C)
Anti-flag M2 antibody (Sigma Aldrich, catalog number: F1804; -20°C)
PBS (Lonza, catalog number: BE17-516F)
NaOH (VWR Chemicals, catalog number: 28244.262)
HCl (Fisher ChemicalTM, catalog number: H/1150/PB17)
Foetal bovine serum (Gibco, catalog number: 10500064; -20°C)
Penicillin-streptomycin (Gibco, catalog number: 15140-122; 4°C)
L-glutamine (Gibco, catalog number: 25030024, 4°C)
HEPES (Sigma-Aldrich, catalog number: H0887)
KCl (Sigma-Aldrich, catalog number: P9541)
NaCl (VWR Chemicals, catalog number: 27810.364)
EDTA (VWR Chemicals, catalog number: 20302.260)
EGTA (Sigma-Aldrich, catalog number: E3889)
NP-40 (IGEPAL® CA-630)( Sigma-Aldrich, catalog number: I3021)
Urea (VWR Chemicals, catalog number: 28877.292)
Sodium deoxycholate (Sigma-Aldrich, catalog number: D6750)
SDS (Sigma-Aldrich, catalog number: 05030)
Precision Plus protein dual color standard (Bio-Rad, catalog number: 1610374)
Gentle RIPA buffer (see Recipes)
Stringent RIPA buffer (see Recipes)
Equipment
Benchtop chilled centrifuge (Thermo Scientific, model: HeraeusTM Fresco 17 or MicroCL 21R)
UV lamp (UVP, model: 95-0007-06)
DNA Shearing Sonicator (Diagenode, model: Bioruptor® Pico or QSonica, model: Q800R1)
Pipette controller (Starlab, model: Starpet Pro E4866-0021)
Tube rotator (Stuart, model: SB3)
Standard orbital shaker (Stuart, model: SSL3)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Isa, N. F., Bensaude, O. and Murphy, S. (2022). Amber Suppression Technology for Mapping Site-specific Viral-host Protein Interactions in Mammalian Cells. Bio-protocol 12(3): e4315. DOI: 10.21769/BioProtoc.4315.
分类
药物发现 > 药物设计
微生物学 > 微生物-宿主相互作用 > 病毒
生物化学 > 蛋白质 > 相互作用 > 交联
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link