- EN - English
- CN - 中文
Heterologous Expression and High Degree Purification of the Restriction Endonuclease SauUSI
限制性内切酶SauUSI的异源表达和高度纯化
发布: 2022年01月05日第12卷第1期 DOI: 10.21769/BioProtoc.4275 浏览次数: 3692
评审: Alba BlesaNingfei AnAnonymous reviewer(s)
Abstract
Mechanisms that target and destroy foreign nucleic acids are major barriers to horizontal gene transfer (HGT) in prokaryotes. Amongst them, restriction-modification (R-M) systems are found in ≥75% of the sequenced genomes in Bacteria and Archaea. Due to their high target sequence specificity and potent nucleolytic activity, R-M systems are used as a paradigm to elucidate the mechanisms of DNA binding and cleavage. Since these enzymes modulate HGT, they are one of the machineries implicated in the ability of a bacterium to gain antibiotic resistance. This protocol provides a detailed purification strategy for the Type IV restriction endonuclease SauUSI from Staphylococcus aureus. This protocol eventually leads to ≥95% purity of protein which can then be used for crystallographic and biochemical purposes.
Graphic abstract:
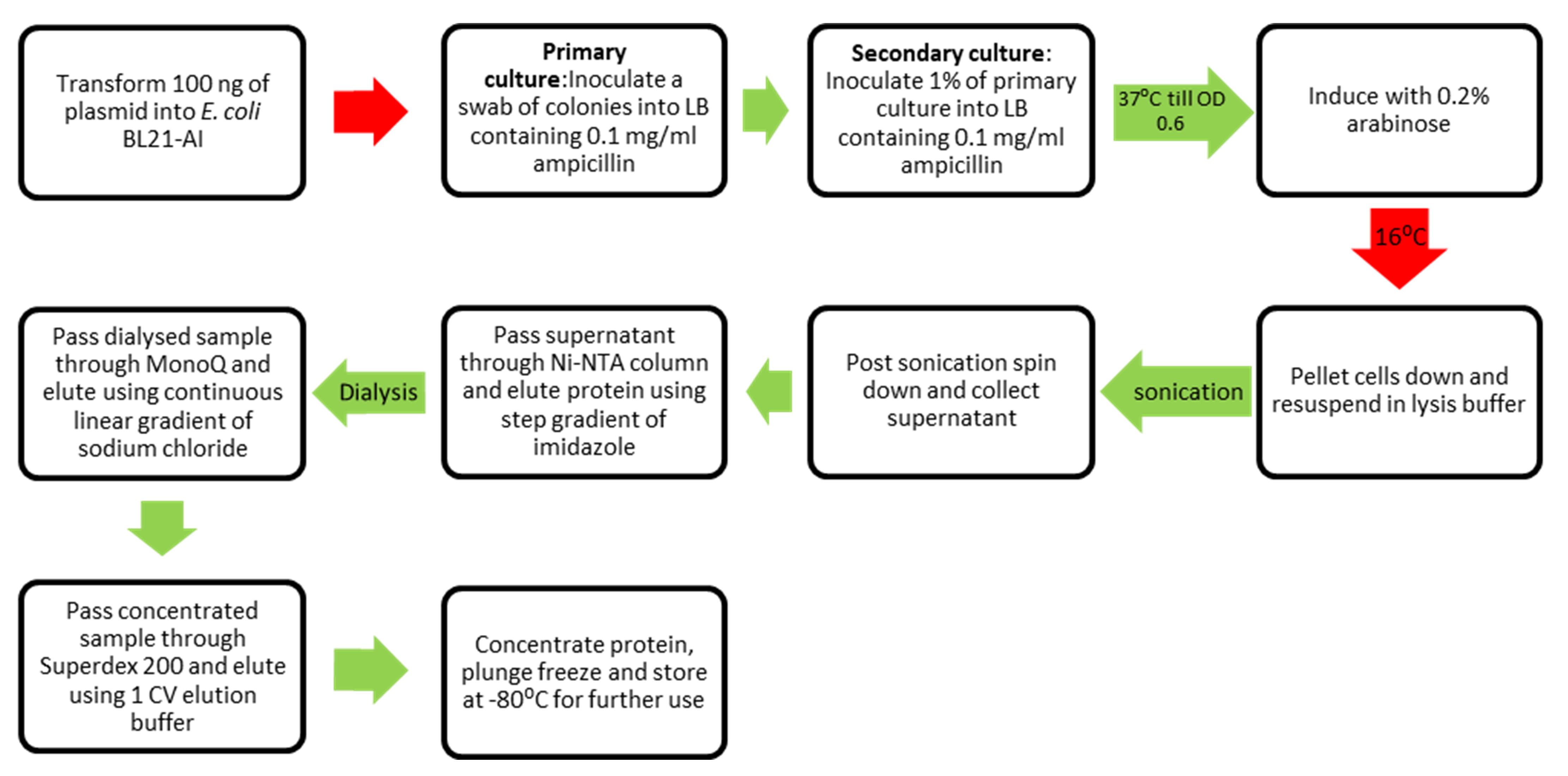
Workflow for purification of SauUSI.
Background
Restriction-modification (R-M) systems function as the innate immune system of bacteria, thus preventing not only infection by bacteriophages but also other potentially harmful mobile genetic elements. There are a variety of bacterial R-M systems that differ in their target site epigenetic status, oligomeric structure, cofactor requirements, and mode of DNA cleavage (Loenen et al., 2014). SauUSI is a Type IV restriction endonuclease present in Staphylococcus aureus that binds specifically to the methylated DNA target sequence 5’-S5mCNGS-3’ (S=C/G and N=A/T/G/C) (Xu et al., 2011). SauUSI has the ability to cleave DNA by two distinct mechanisms; they are: a) a highly efficient convergence-based cleavage mechanism when there are two or more target sequences, and b) a lesser efficient switch-based mechanism on DNA with only one target sequence (Tumuluri et al., 2021). Studies on the cleavage activity of SauUSI highlight its role in preventing the acquisition of exogenous genetic elements that have the ability to transform methicillin-resistant Staphylococcus aureus (MRSA) into vancomycin-resistant Staphylococcus aureus (VRSA) (Corvaglia et al., 2010; Monk et al., 2012; Tumuluri et al., 2021).
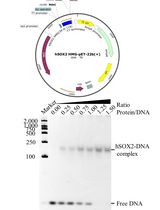
For the in-vitro biochemical characterization and crystallographic studies, SauUSI was purified post expression in the heterologous Escherichia coli BL21-AI cells. For this purpose, the sauUSIR gene was cloned into a pHis17 (ampicillin resistant) vector (Kunzelmann and Webb, 2009), between the NdeI (upstream) and BamHI (downstream) restriction sites (Figure 1), using a restriction-free (RF) cloning strategy (van den Ent and Löwe, 2006; Tumuluri et al., 2021). The RF cloning primarily consisted of two sequential polymerase chain reactions (PCRs): the first reaction was to amplify the sauUSIR gene with flanking regions complementary to the pHis17 vector, and the second reaction was an extension PCR to incorporate the amplified sauUSIR gene into the vector. The reaction was treated with DpnI to cleave the parent template, and subsequentially transformed into Escherichia coli NEB Turbo® cells. The presence of the sauUSIR gene was confirmed by DNA sequencing.
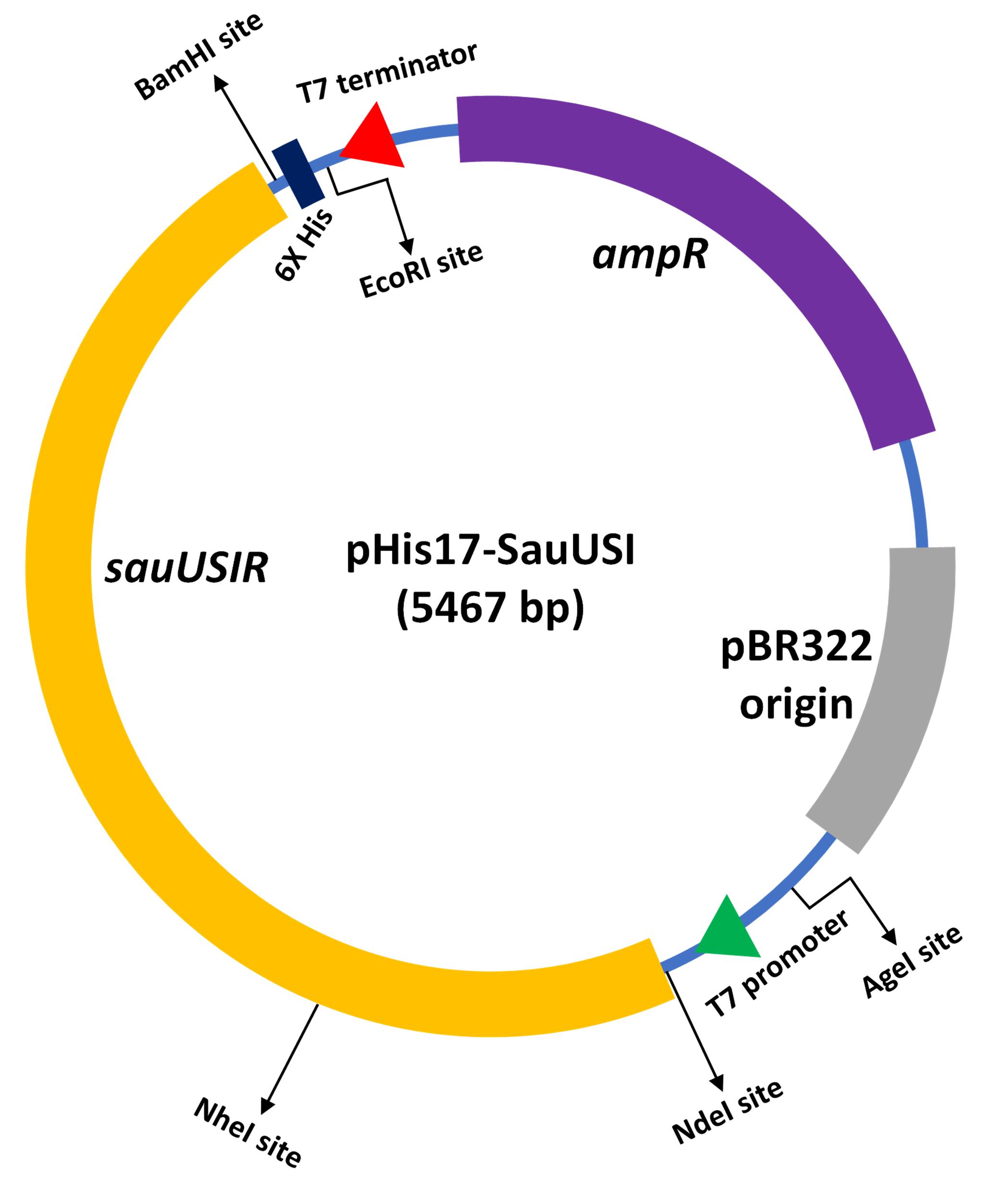
Figure 1. Cartoon representation of the expression vector integrated with sauUSIR gene. The sauUSIR gene is 2859 bp and codes of a 953 amino acid single polypeptide protein. This gene is integrated between NdeI and BamHI sites in a pHis 17 vector thus providing a C-terminal 6× Histidine tag for ease of purification.
Materials and Reagents
100 mm Petri dishes (HiMedia, catalog number: PW1305-1x100NO)
50 mL beaker (Borosil)
200 mL conical flask (Borosil)
2000 mL culture flask (Borosil)
50 mL centrifuge tubes (Eppendorf)
15 mL centrifuge tubes (Eppendorf)
1.5 mL fraction tubes (Eppendorf)
0.5 mL fraction tubes (Eppendorf)
SnakeSkinTM dialysis tubing 10000 MWCO (ThermoFisher, catalog number: 68100)
0.2 µm syringe filter (WhatmanTM, catalog number: 9914-2502)
0.2 µm filter membranes (Millipore, catalog number: GVWP04700)
Whatman filter paper (No. 1) (Filtration laboratory, catalog number: T-06648-13)
Lysogeny broth (LB) (HiMedia, catalog number: M1245-1KG)
Luria-Bertani (LB) Agar (HiMedia, catalog number: M1151-1KG)
Ampicillin (Sigma-Aldrich, catalog number: A9518-25G)
Arabinose (SRL, catalog number: 52392)
Sodium Chloride (NaCl) (Sigma-Aldrich, catalog number: S9888-1KG)
Tris(hydroxymethyl)aminomethane (Tris-Base) (Sigma-Aldrich, catalog number: 252859-500G)
Hydrochloric acid (HCl) for molecular biology (Qualigens, catalog number: Q29505)
Imidazole (Sigma-Aldrich, catalog number: I2399-500G)
Magnesium Chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266-100G)
3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) (Sigma-Aldrich, catalog number: 226947-5G)
Glycerol for molecular biology (Sigma-Aldrich, catalog number: 356352-1L-M)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: ED-500G)
1,4-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 10708984001)
Glycine (Sigma-Aldrich, catalog number: G8898-1KG)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771-100G)
Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: A3678-25G)
Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281-25ML)
Acrylamide (Sigma-Aldrich, catalog number: A8887-500G)
Bis-acrylamide (Sigma-Aldrich, catalog number: 146072-500G)
Bio-Rad Precision Plus ProteinTM Standards (Bio-Rad, catalog number: 1610374)
2× Laemmli sample buffer (Bio-Rad, catalog number: 1610737EDU)
Coomassie Brilliant Blue (R-250) (Sigma-Aldrich, catalog number: 1125530025)
Ethanol for molecular biology
Glacial Acetic acid for molecular biology (Qualigens, catalog number: Q21055)
Liquid nitrogen
Stock solutions (see Recipes)
20% (w/v) Arabinose
5 M NaCl
2 M Tris-HCl (pH = 8)
2 M Imidazole
4% (w/v) CHAPS
2 M MgCl2
0.5 M EDTA (pH = 8)
1 M DTT
Buffers for purification (see Recipes)
Lysis buffer
Buffers for affinity (Ni-NTA) chromatography (Buffer A and Buffer B)
Buffer for Dialysis (Buffer B0)
Buffers for anion exchange chromatography (Buffer B50 and Buffer B1000)
Buffer for size exclusion chromatography (Buffer B100)
SDS-PAGE (see Recipes)
10× stock of TGS (Tris base, glycine, and SDS) running buffer
Coomassie staining solution
Coomassie destaining solution
Solutions required for Gel mix
10% resolving gel
5% stacking gel
Equipment
Thermoblock (Eppendorf ThermoMixer C, catalog number: 5382000015)
Shaker incubator (ThermoScientific, model: MXQ 6000)
Avanti J26S-XP centrifuge (Beckman Coulter, model: Avanti J26S-XP, Rotor: JLA-9.1000)
Optima-L-100K ultracentrifuge (Beckman Coulter, model: Optima L-100K, Rotor: Type 45 Ti)
GE AKTAprime plus FPLC system (GMI, catalog number: 8149-30-0004)
Bio-Rad NGCTM chromatography system (Bio-Rad, catalog number: 788001)
Vacuum filter assembly (Millipore, model: XX1014720)
Sonicator (Sonics-Vibra cell) (Sonics, model: VCX 130)
HisTrapTM HP 5 mL Ni-NTA column (GE, catalog number: 17524802)
MonoQTM 10/100 GL column (GE, catalog number: 17516701)
SuperloopTM (50 mL) (GE, catalog number: 18111382)
Superdex 200TM 16/600 pg column (GE, catalog number: GE28-9893-35)
SDS-PAGE apparatus (Bio-Rad, catalog number: 1658001FC)
VivaspinR 10000 MWCO centrifugal concentrator (Sartorius, catalog number: Z614602-12EA)
Software
Microsoft Excel
GraphPad Prism
ImageJ
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Tumuluri, V. S. and Saikrishnan, K. (2022). Heterologous Expression and High Degree Purification of the Restriction Endonuclease SauUSI. Bio-protocol 12(1): e4275. DOI: 10.21769/BioProtoc.4275.
分类
生物化学 > 蛋白质 > 表达
分子生物学 > 蛋白质 > 表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link