- EN - English
- CN - 中文
Selection of Vaccinia Virus Recombinants Using CRISPR/Cas9
使用 CRISPR/Cas9 筛选牛痘病毒重组体
发布: 2021年12月20日第11卷第24期 DOI: 10.21769/BioProtoc.4270 浏览次数: 3404
评审: ASWAD KHADILKARBruno HernaezIsmar HagaJonas Albarnaz

相关实验方案
利用小鼠Phospho-RTK Array试剂盒无偏筛选肿瘤提取物中激活受体酪氨酸激酶
Julian Naipauer [...] Enrique A. Mesri
2019年04月20日 5671 阅读
Abstract
The engineering of poxvirus genomes is fundamental to primary and applied virology research. Indeed, recombinant poxviruses form the basis for many novel vaccines and virotherapies but producing and purifying these viruses can be arduous. In recent years, CRISPR/Cas9 has become the favoured approach for genome manipulation due to its speed and high success rate. However, recent data suggests poxvirus genomes are not repaired well following Cas9 cleavage. As a result, CRISPR/Cas9 is inefficient as an editing tool, but very effective as a programmable selection agent. Here, we describe protocols for the generation and enrichment of recombinant vaccinia viruses using targeted Cas9 as a selection tool. This novel use of Cas9 is a simple addition to current homologous recombination-based methods that are widespread in the field, facilitating implementation in laboratories already working with poxviruses. This is also the first method that allows for isolation of new vaccinia viruses in less than a fortnight, without the need to incorporate a marker gene or manipulation of large poxvirus genomes in vitro and reactivation with helper viruses. Whilst this protocol describes applications for laboratory strains of vaccinia virus, it should be readily adaptable to other poxviruses.
Graphic abstract:
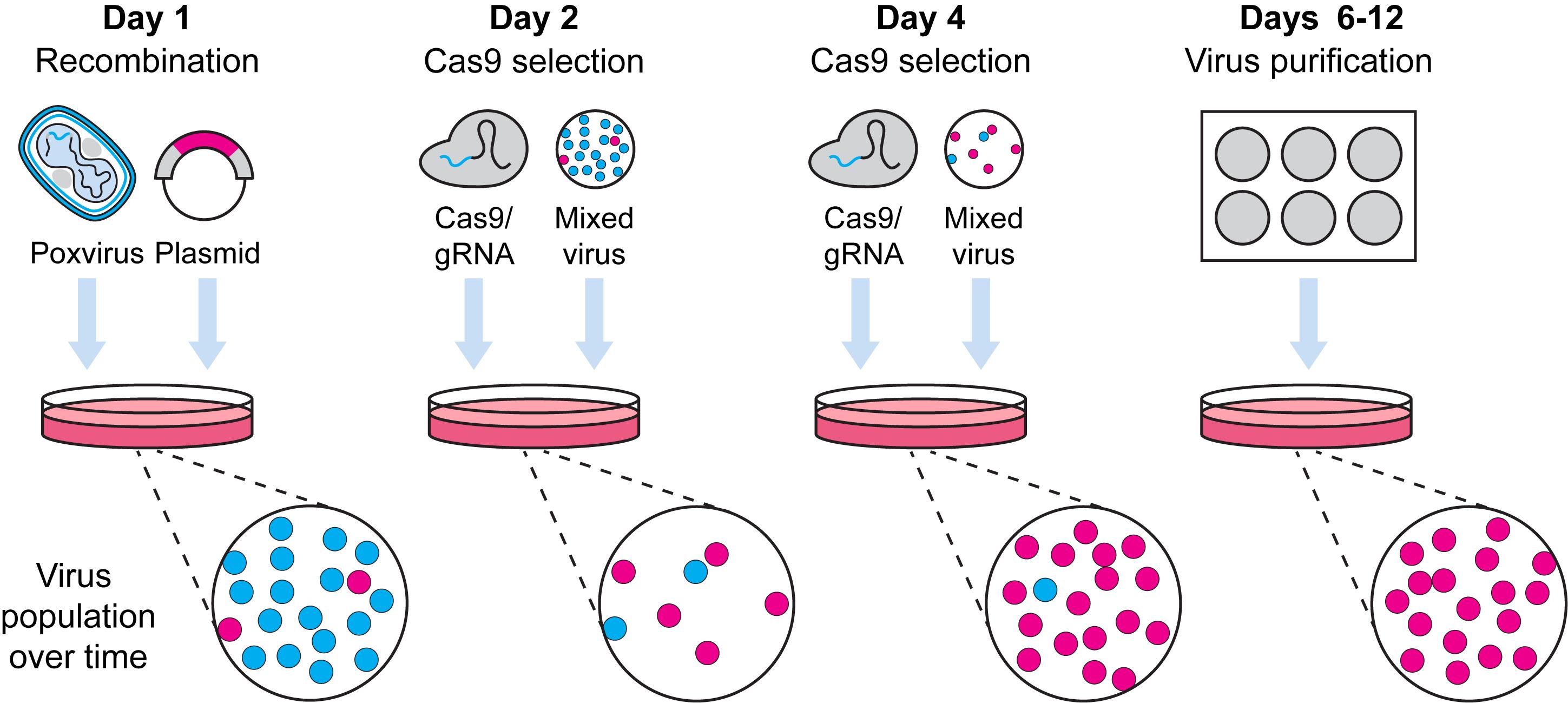
Pipeline for Cas9 selection of recombinant poxviruses.
Background
Poxviruses are important research viruses, both in the context of infection models and as vectors for many virotherapeutics. A significant part of their usefulness lies in their genomes, which are amenable to substantial manipulation (Smith and Moss, 1983). This allows researchers to incorporate foreign genes, such as those required for immunomodulation or markers for virus tracking, to remove viral genes, as is often required for pathogenesis studies, and to introduce point-mutations of interest. However, existing processes for manipulating poxvirus genomes, including homologous recombination or transient-dominant selection, can be inefficient and time-consuming.
CRISPR/Cas9 editing has wide applications that includes modifying viruses with double-stranded DNA genomes (Russell et al., 2015; Yuen et al., 2015; King and Munger, 2019). These methods can be thought of as using a two-step, “cut and paste” approach for DNA manipulation – Cas9 nuclease is first directed to target sites by guide RNA molecules (gRNAs) and forms double-stranded DNA breaks, then cellular enzymes repair these sites introducing mutations. In the absence of repair templates, DNA repair proceeds via non-homologous end joining, which is error-prone and can lead to the incorporation of random indels. Alternatively, when a repair template having ends that share identity with sequences that flank the target site is present, specific genetic changes can be made. However, for poxviruses that replicate in the cell cytoplasm, repair of Cas9 cleavage is very inefficient (Gowripalan et al., 2020). This has a profound antiviral effect and allows the repurposing of targeted Cas9 as a selection tool rather than as one that generates gene edits.
Here, we describe a method for Cas9 selection of recombinant vaccinia viruses (VACV) (Figure 1). To make a complete protocol, we start by sharing our method for generating VACV by homologous recombination. There are many other variations of this method and any of these should be equally effective. This is followed by our method for using Cas9/gRNA complexes to target parent virus genomes, removing these and leading to enrichment of the desired recombinant. Multiple rounds of selection further increase the selective pressure and, over time, we observe recombinant viruses becoming the dominant population. The process is robust, allowing for generation of both marker-containing and marker-free viruses. Our protocol is for laboratory strains of VACV but should be adaptable to other poxviruses, the proviso being that a cell type that is permissive for the virus and is highly transfectable can be found for selection. It should be noted that the requirement for high transfection efficiency is greater for Cas9 selection than for the initial recombination.
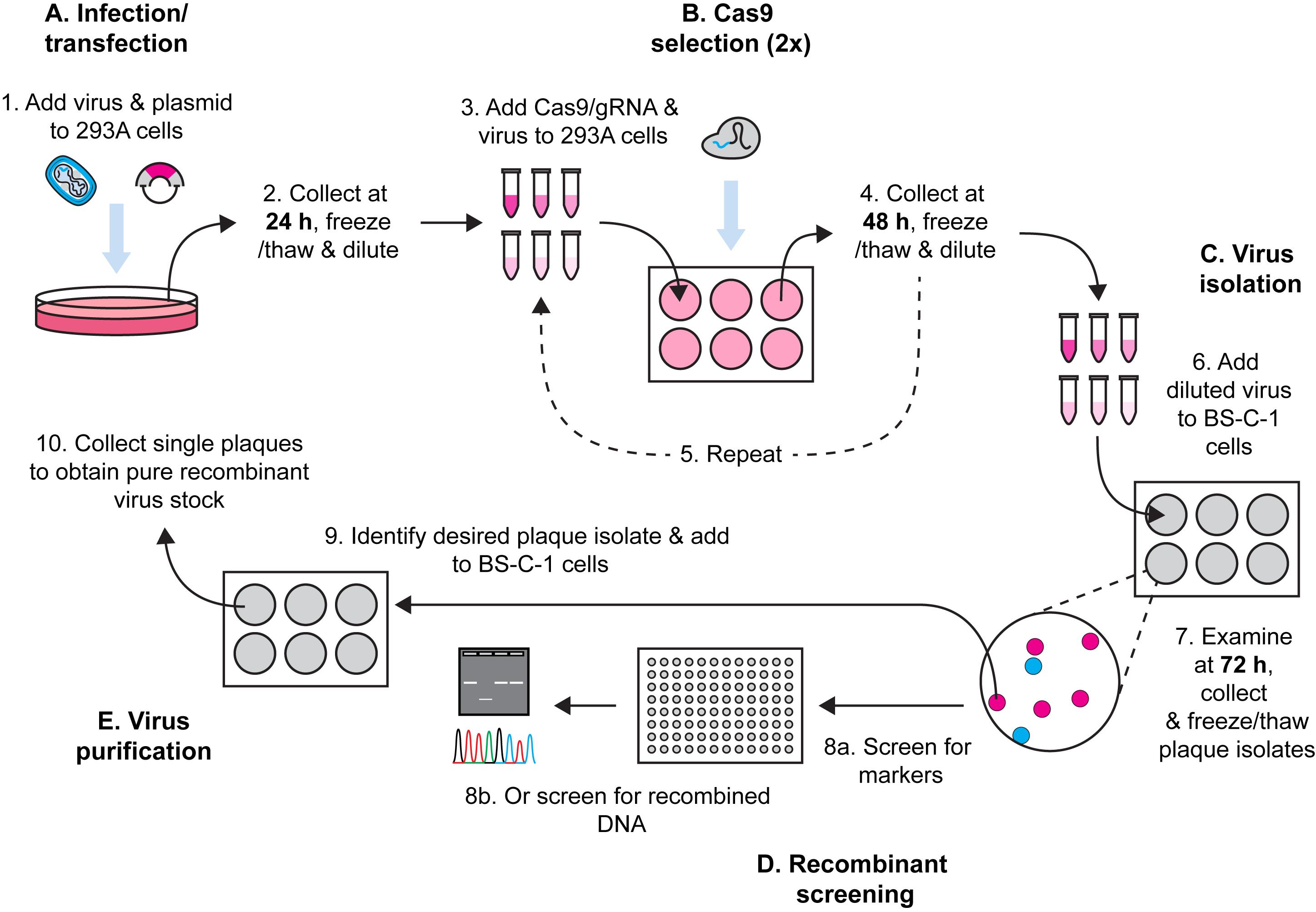
Figure 1. Workflow for generation of recombinant VACVs using Cas9 selection.
Materials and Reagents
6-well plates, flat bottom (Corning® Costar, catalog number: CLS3516)
96-well plates, flat bottom (Corning® Costar, catalog number: CLS3595)
Pipette tips
Serological pipettes
2.0 ml cryogenic vials (Corning®, catalog number: 430659)
2.0 ml Sarstedt tubes (Sarstedt, catalog number: 72.693.005)
5 ml Sarstedt tubes (Sarstedt, catalog number: 60.9921.532)
30 ml Sarstedt tubes (Sarstedt, catalog number: 60.9922.216)
8-strip Sarstedt PCR tubes (Sarstedt, catalog number: 72.991.002)
Dulbecco’s Modified Eagle’s Medium – high glucose (Millipore Sigma, catalog number: D5671)
Dulbecco’s Modified Eagle’s Medium – high glucose, no glutamine, no phenol red (Thermo Fisher Scientific, catalog number: 31053028)
L-Glutamine (200 mM) (Thermo Fisher Scientific, catalog number: 25030081)
Fetal bovine serum (FBS) (Millipore Sigma, catalog number: 12003C-500 mL)
Trypsin-EDTA (0.5%), no phenol red (Thermo Fisher Scientific, catalog number: 15400054)
Dulbecco’s Phosphate Buffered Saline (Millipore Sigma, catalog number: D8537)
LipofectamineTM 2000 (Thermo Fisher Scientific, catalog number: 11668019)
293A (Human embryonic kidney epithelial) cells (Thermo Fisher Scientific, catalog number: R70507)
BS-C-1 (African Green monkey kidney epithelial) cells (ATCC, catalog number: CCL-26)
VACV WR (NIH TC-adapted; ATCC VR1354), often referred to as Western Reserve (source was B. Moss, National Institutes of Health). Other replication competent strains should behave similarly.
VACV mCherry (derivative of VACV WR) (Gowripalan et al., 2020)
Cas9 nuclease, S. pyogenes with 10× Cas9 nuclease reaction buffer (New England Biolabs, catalog number: M0386M)
gRNAs purchased as Ultramer® RNA Oligos, stored at -80°C (Integrated DNA Technologies, see Note 1)
Plasmids for recombination synthesized prior to experimentation and stored at -20°C (see Note 2)
NucleoSpin Plasmid Mini Kit (Machery-Nagel, catalog number: 740588.250)
Carboxymethyl cellulose (CMC) sodium salt (Millipore Sigma, catalog number: C4888-500G)
100% ethanol (Ajax AR Grade, catalog number: AJA214)
Sharpie® Ultra Fine Permanent Marker (Sharpie, catalog number: 37001)
Taq DNA polymerase with ThermoPol® Buffer (New England Biolabs, catalog number: M0267X)
Proteinase K (Fungal) (Invitrogen, catalog number: 25530-015)
Stocks are typically made at 10 mg/ml in 50 mM Tris pH 8 and stored at -20°C and dilutions are made in nuclease-free water.dNTP Mix (Bioline, catalog number: BIO39053)
PCR primers purchased as Custom DNA oligos and stored at -20°C (Integrated DNA Technologies)
Gel loading dye, Purple (6×) (New England Biolabs, catalog number: B7024S)
UltraPureTM Agarose (Thermo Fisher Scientific, catalog number: 16500500)
SYBRTM Safe DNA Gel Stain (Thermo Fisher Scientific, catalog number: S33102)
D0 media (see Recipes)
D2 media (see Recipes)
D2 media – no phenol red (see Recipes)
D10 media (see Recipes)
D10 media – no phenol red (see Recipes)
4% CMC/D2 – no phenol red (see Recipes)
Equipment
Class 2 biosafety cabinet (Euroclone, model: Safemate Vision 1.2 ABC)
Incubator with CO2 (Thermo Fisher Scientific, model: Heracell 150i CO2)
-80°C freezer (Thermo Fisher Scientific, model: Forma 705 -80°C)
Branson Analog Sonifier S-450 Sonicator (Branson, model: S-450; 102 with 3” cup)
Olympus CKX53 inverted microscope (Olympus, model: N5731900)
Dry-block heater with digital control (Major Science, catalog number: MD-02N)
Veriti® PCR thermocycler (Applied Biosystems, catalog number: 4375786)
Table-top centrifuge (Eppendorf, model: Centrifuge 5424)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Gowripalan, A., Smith, S. A. and Tscharke, D. C. (2021). Selection of Vaccinia Virus Recombinants Using CRISPR/Cas9. Bio-protocol 11(24): e4270. DOI: 10.21769/BioProtoc.4270.
分类
微生物学 > 微生物遗传学 > DNA
微生物学 > 体内实验模型 > 病毒
分子生物学 > DNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












