- EN - English
- CN - 中文
Copper Based Site-directed Spin Labeling of Proteins for Use in Pulsed and Continuous Wave EPR Spectroscopy
用于脉冲和连续波 EPR 光谱的基于铜的蛋白质定点自旋标记
发布: 2021年12月20日第11卷第24期 DOI: 10.21769/BioProtoc.4258 浏览次数: 3739
评审: Manjula MummadisettiBela BodeAnonymous reviewer(s)
Abstract
Site-directed spin labeling in conjunction with electron paramagnetic resonance (EPR) is an attractive approach to measure residue specific dynamics and point-to-point distance distributions in a biomolecule. Here, we focus on the labeling of proteins with a Cu(II)-nitrilotriacetic acid (NTA) complex, by exploiting two strategically placed histidine residues (called the dHis motif). This labeling strategy has emerged as a means to overcome key limitations of many spin labels. Through utilizing the dHis motif, Cu(II)NTA rigidly binds to a protein without depending on cysteine residues. This protocol outlines three major points: the synthesis of the Cu(II)NTA complex; the measurement of continuous wave and pulsed EPR spectra, to verify a successful synthesis, as well as successful protein labeling; and utilizing Cu(II)NTA labeled proteins, to measure distance constraints and backbone dynamics. In doing so, EPR measurements are less influenced by sidechain motion, which influences the breadth of the measured distance distributions between two spins, as well as the measured residue-specific dynamics. More broadly, such EPR-based distance measurements provide unique structural constraints for integrative structural biophysics and complement traditional biophysical techniques, such as NMR, cryo-EM, FRET, and crystallography.
Graphic abstract:
Monitoring the success of Cu(II)NTA labeling.
Background
Electron paramagnetic resonance (EPR) holds a unique position in structural biology as it focuses on residue-specific information, rather than the global structure that other techniques strive for. The first focus of EPR employs pulsed strategies to provide nanometer-scale distance measurements between two or more spin labels. While multiple pulsed techniques exist, the most common method, which is double electron-electron resonance (DEER), works by flipping the spin of one electron spin and measuring its influence on another nearby spin (Pannier et al., 2000). This information can be used incisively to measure induced conformational changes, locate binding sites, and establish quaternary structure, as well as the relative positioning of two biomolecules (Park et al., 2006; Gaffney et al., 2012; Jeschke, 2012). The other application of EPR in structural biology focuses on the relationship of protein function to site-specific dynamics. The EPR lineshape is sensitive to the tumbling rate of the paramagnetic species. Therefore, this lineshape provides information on residue specific motional rates (Hubbell et al., 1998).
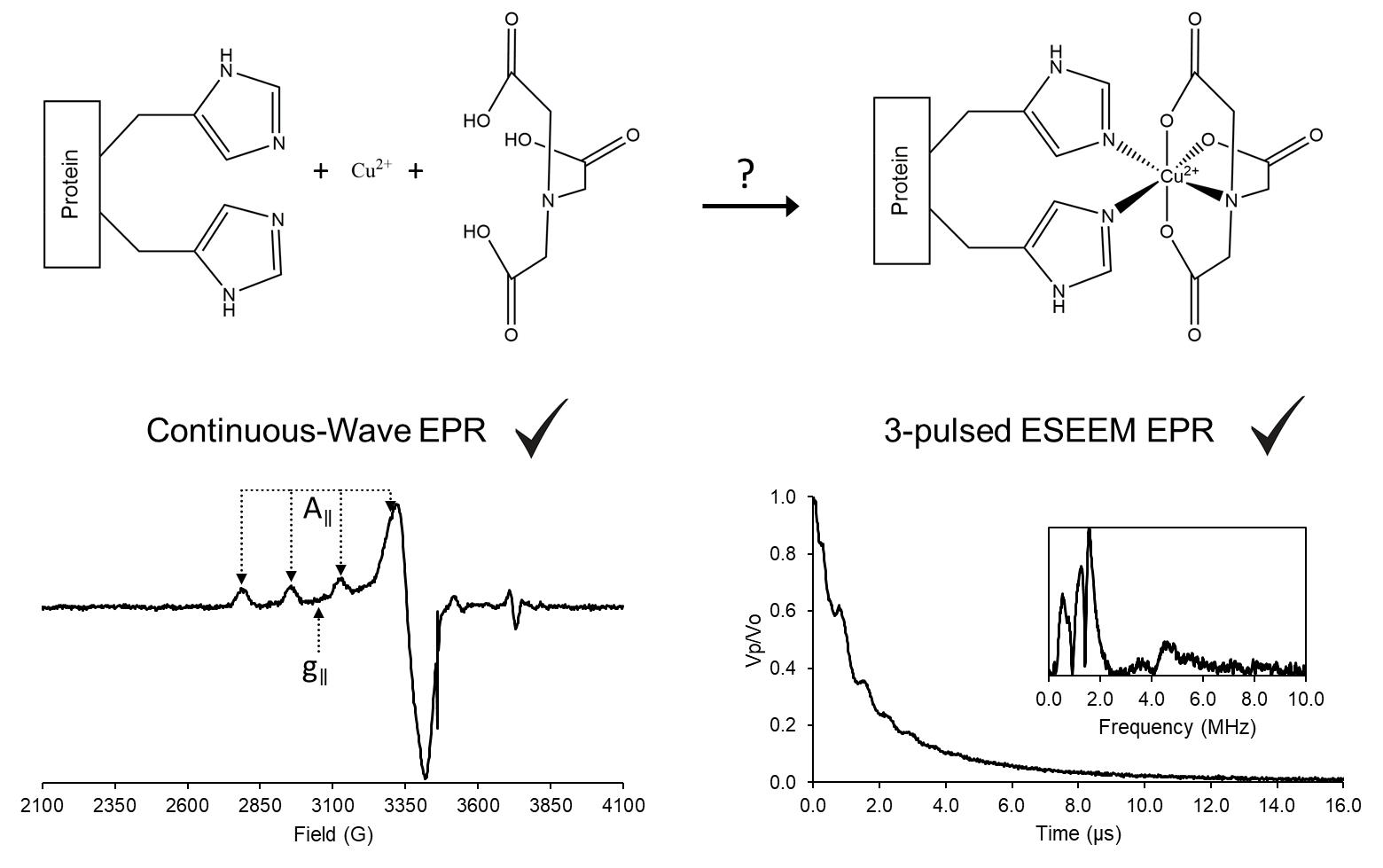
In order to use EPR, spin labeling is a key component as most biomolecules are diamagnetic, i.e., EPR inactive (Todd et al., 1989). In fact, spin labeling enables selective measurements with minimal to no background interference. Most spin labels utilize cysteine residues as attachment points to the protein (Klug et al., 2021). However, labeling a specific cysteine may require other native cysteines to be mutated out, which can be problematic in many cases (Poole, 2015; Qiu et al., 2015). Additionally, the intrinsic flexibility of most spin labels can influence the measured distance distribution between two spins, as well as the measured side-chain dynamics (Klose et al., 2012). On the other hand, site-directed Cu(II) labeling to the double histidine motif (dHis) overcomes these limitations. This labeling is reliant on Cu(II) binding to two strategically placed histidine residues of a protein: i and i+2 for β-sheets, and for α-helices i and i+4. Furthermore, the use of the Cu(II)NTA complex enhances the selectively to the dHis motif, while minimizing native or non-specific Cu(II) binding (Ghosh et al., 2018). The bi-pedal attachment of Cu(II)NTA to two His residues leads to distance distributions that can be up to 5-times narrower than traditional nitroxide labels (Cunningham et al., 2015). Correspondingly, the dHis motif has been exploited to study subtle conformational changes that cannot be resolved with standard nitroxide radicals (Sameach et al., 2019), locate native metal binding sites with fewer distance constraints (Gamble Jarvi et al., 2019), and directly analyze backbone dynamics (Singewald et al., 2020). On the other hand, this technique has not been thoroughly explored for buried regions in a protein or for membrane proteins. This protocol aims to outline Cu(II) labeling of proteins and how to monitor successful labeling.
Materials and Reagents
Mechanical micropipettes (Eppendorf, catalog number: 2231302001)
Matching pipette tips for the mechanical micropipettes (Fisher Scientific, catalog numbers: 02-707-507, 02-707-450, 02-717-133)
1.5 ml Microcentrifuge tube (Eppendorf, catalog number: 022363204)
Conical tubes, 15 ml (Thermo Fisher Scientific, catalog number: 339650)
Quartz EPR tubes, 3 mm I.D. × 4 mm O.D. × 250 mm length (Sigma-Aldrich, catalog number: Z566535)
Quartz capillary tubes, 0.8 mm I.D. × 1 mm O.D. × 100 mm length (VitroCom, catalog number: CV8010-Q-100)
Capillary tube sealant (Fisher Scientific, catalog number: 02-678)
PTFE tape, medium density, 12.7 mm × 13.21 mm (Anti-Seize Technology, catalog number: 16035)
Dewar shielded vacuum flasks for freezing in liquid nitrogen, 350 ml (Pope Scientific, catalog number: 8600-0099)
Low form Dewar flask, 275 ml (Cole-Parmer, catalog number: EW-03771-21)
Metal wire, approximately 50 cm
Brass cylinder, 20 mm diameter × 30 mm length, with a blind hole on one end, 7 mm diameter × 25 mm length
Gas regulator (Uniweld, catalog number: RP3E)
Tygon E-3603 tubing, 5/16” I.D. × 7/16” O.D. (Cole-Parmer, catalog number: EW-07407-83)
Safety goggles (Fisher Scientific, catalog number: 12-888-305)
Cryogen protecting gloves (Grainger, catalog number: 2AEY6)
Deionized water (Fisher Scientific, catalog number: 21-520-113)
Copper sulfate pentahydrate (Spectrum Chemical, catalog number: C1431)
Hydrochloric acid, needed to prepare 1 M HCl solution for pH adjustment (Fisher Scientific, catalog number: 02003059)
Sodium hydroxide pellets, needed to prepare 1 M NaOH solution for pH adjustment (Fisher Scientific, catalog number: 56-753-0250GM)
Calibration solutions (pH = 4.00, 7.00, 10.00) (Fisher Scientific, catalog numbers: SB101-500, SB107-500, SB115-500)
pH electrode storage solution (Hanna Instruments, catalog number: HI70300)
pH electrode fill solution (Hanna Instruments, catalog number: HI7082)
pH electrode storage cap (Hanna Instruments, catalog number: HI740200)
Sodium chloride (Fisher Scientific, catalog number: BP3581)
MOPS (Fisher Scientific, catalog number: BP358)
Nitrilotriacetic acid disodium, NTA (Sigma-Aldrich, catalog number: N0128)
Glycerol (Sigma-Aldrich, catalog number: G7757)
Ficoll PM 70 (Fisher Scientific, catalog number: 45002020)
MAP-Pro Propylene/propane gas (Bernzomatic, catalog number: MG9)
Liquid nitrogen for CW-EPR and ESEEM experiments
Liquid helium for DEER/PELDOR experiments
Protein with one or more dHis mutations
100 mM MOPS Buffer (see Recipes)
Note: Deuterated solvents may be used to increase the Tm relaxation time for experiments that require longer evolution times (Casto et al., 2021).
Equipment
Analytical balance (Denver Instruments, model: M-220D)
Vortex-Genie 2 (Scientific Industries, catalog number: SI-0236)
pH electrode (Hanna Instruments, catalog number: HI1332B)
pH meter, accumet AB150 (Fisher Scientific, catalog number: 13-636-AB150A)
Refrigerator 4°C
Freezer -20°C and -80°C
Spectrophotometer to determine protein concentration (Thermo Scientific, model: NanoDrop 2000)
Bruker ElexSys E680 CW/FT X-band spectrometer
Bruker ER4118-MD5 resonator
Bruker ER4122 SHQE-W1 high resolution resonator
Oxford ITC503 temperature controller
Oxford LLT 650 low loss transfer tube
Oxford CF935 dynamic continuous-flow cryostat
Software
MATLAB Version R2020b or later (The MathWorks Inc.) https://www.mathworks.com/
EasySpin toolbox for MATLAB, Version 5.2.30, available for free (Stoll and Schweiger, 2006) https://www.easyspin.org/
Bruker Xepr software provided with EPR spectrometer
NanoDrop 2000 program provided with spectrophotometer
A program to do NLSL.MOMD analysis, available for free (Budil et al., 1996) https://www.acert.cornell.edu/index_files/acert_ftp_links.php
Xepr ESEEM program and example scripts for use in EasySpin and NLSL are available at https://github.com/SaxenaLab/Copper
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Singewald, K., Wilkinson, J. A. and Saxena, S. (2021). Copper Based Site-directed Spin Labeling of Proteins for Use in Pulsed and Continuous Wave EPR Spectroscopy. Bio-protocol 11(24): e4258. DOI: 10.21769/BioProtoc.4258.
分类
生物物理学 > EPR波谱
生物化学 > 蛋白质 > 标记
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link