- EN - English
- CN - 中文
A Combinatorial in-silico, in-vitro and in-vivo Approach to Quantitatively Study Peptide Induced MHC Stability
结合硅、体外和体内方法定量研究多肽诱导的MHC稳定性
发布: 2021年12月20日第11卷第24期 DOI: 10.21769/BioProtoc.4255 浏览次数: 4471
评审: Luis Alberto Sánchez VargasMarie BoutetRAMESH KUDIRA
Abstract
Here, we describe a combinatorial approach in reverse vaccinology to identify immunogenic class I major histocompatibility complex (MHC) displayed epitopes derived from a morbillivirus named pestes des petits ruminants (PPRV). The protocol describes an in silico prediction of immunogenic epitopes using an IEDB tool. The predicted peptides were further analysed by molecular docking with mouse class I MHC (H-2Kb), to assess their binding affinity, and their immunogenicity was validated, using acellular and cellular assays. Finally, an enumeration of the expanded PPRV-specific CD8+ T cells in infected or immunized mice against the immunogenic peptides was performed ex vivo. Synthetic peptide derivatives from different structural and non-structural proteins of PPRV were used to measure the extent of stabilized H2-Kb, using an ELISA based acellular assay and TAP deficient RMA/s cells. Fluorescently labelled H2-Kb-tetramers were generated by displacing a UV photocleavable conditional ligand with the PPRV-peptides. The resulting reagents were used to identify and enumerate virus-specific CD8+ T cells in immunized or PPRV-infected mice. The combinatorial approach described here could be used to identify immunogenic epitopes of any pathogen, autoantigens, as well as cancer antigens.
Graphic abstract:
Figure 1. General schematic to identify immunogenic peptides and their stabilization on MHC I molecule.
Background
Major histocompatibility complex (MHC) molecules loaded with peptides are displayed on the cell surface to engage T cells via their expressed TCRs. While all nucleated cells express class I MHC molecules, professional APCs such as dendritic cells, macrophages, and B cells additionally express high level of class II MHC molecules to activate T cells (Wieczorek et al., 2017). Formation of assembled class I and II MHC molecules requires binding of the proteolytically generated peptides with specific positioning of anchor residues. Endogenously synthesized proteins or those acquired exogenously by APCs are processed in distinct subcellular compartments to generate peptides that are presented by class I MHC molecules at the cell surface (Princiotta et al., 2003). Endogenous proteins are primarily processed by proteasomal machinery, and peptides thus generated are translocated from cytosol to endoplasmic reticulum (ER) mainly by the transporter associated with antigen processing (TAP) molecules (Vyas et al., 2008). Nascent class I MHC molecules in the ER associate with calreticulin, tapasin, and ERp57 to generate a peptide loading complex (PLC) that facilitates loading of peptides. Then, the assembled complexes traverse to the cell surface. Therefore, TAP deficiency limits the entry of peptides into ER, leading to a reduced surface expression of class I MHC molecules. The heterotrimeric complex formed by heavy chain (HC) of class I MHC molecule, peptide, and β2 microglobulin (β2m) provides the central anchorage signal to engage specific TCR expressing cytotoxic T lymphocytes (CTLs). Additionally, the co-stimulatory receptor-ligand pair (B7.1/7.2 and CD28) by the engaged APCs and T cells helps to efficiently activate CTLs, which are then enormously expanded in the presence of generated cytokine milieu (De Bruijn et al., 1991; La Gruta et al., 2018). Such cells scan infected or transformed cells for cytolysis. Therefore, identifying pathogen derived peptides constitutes a critical step in understanding T cell dynamics following infection or immunization. To validate the immunogenicity of the predicted peptides, fluorescently labelled H-2Kb/peptide complexes were generated and used to detect PPRV-peptides specific CD8+ T cells in immunized or the PPRV-infected mice.
In this protocol, we describe assays to ascertain peptide induced class I MHC stabilization (Wieczorek et al., 2017). A combinatorial approach involves in silico, in vitro and in vivo analysis to predict immunogenic PPRV-peptides that can be displayed by class I MHC molecules of mice (H-2Kb). Finally, a throughput approach was used to enumerate PPRV-specific CD8+ T cells in the immunized or infected mice. For identifying such peptides, we collected the complete amino acid sequences of PPRV proteins from online NCBI and Uniprot databases to predict class I MHC restricted epitopes using online tools such as IEDB, SYFPEITHI (Figure 1). High affinity epitopes were manually selected based on their hydrophobic residues at carboxy terminus, as the latter are known to favor peptide-MHC class I interaction (Pettersen et al., 2004; Chen et al., 2015). Furthermore, the selected high affinity peptides were subjected to a blind docking analysis with H-2Kb, whereby the top ranked peptides were selected (Vita et al., 2015 and 2019; Zhou et al., 2018). To study the presentation of such peptides in context to H-2Kb, we used TAP deficient RMA/S cells that inherently express low levels of surface class I MHC molecules. RMA/s cells can synthesize heavy chains of class I MHC molecules and β2m, but fail to efficiently assemble these molecules due to the limited availability of processed peptide in the ER (Esquivel et al., 1992; De Silva et al., 1999). This leads to compromised loading of class I MHC molecules with antigenic peptides. Empty class I MHC molecules can, nonetheless, be transported to the cell surface by vesicular transport from the ER (De Bruijn et al., 1991; Esquivel et al., 1992). Empty class I MHC molecules are poorly stabilized at the cell surface and hence lower expression levels are evident. Exogenously added antigenic peptides to RMA/S cells can be loaded to class I MHC molecules by predominantly two mechanisms. First, the exogenous peptides can bind directly to the empty class I MHC molecules at the cell surface (Schumacher et al., 1990). Second, the peptides are internalized by such cells, loaded onto the peptide binding grove of heavy chain (HC) of class I MHC molecule utilizing the vacuolar pathway to generate stable heterotrimeric complexes involving β2m, and subsequently the complexes are efficiently displayed on the cell surface (Figure 2) (Esquivel et al., 1992; De Silva et al., 1999). Fine details of such processes are still to be better defined. The binding ability of anchor residues of the peptide with MHC molecules dictates the stability of such complexes (Garstka et al., 2015). We showed that stable class I MHC molecules were expressed on the cell surface when immunogenic peptides derived from PPRV were added to RMA/s cells, and such molecules were detected by anti-H-2Kb antibody using flow cytometry (Sharma et al., 2021). We also generated stable H-2Kb monomers using a photocleavable conditional ligand, (FAPG[Anp]YPAL), modified derivative from the nucleoprotein of Sendai E virus, which was described earlier byRodenko et al. (2006). Incubation of predicted PPRV-peptides in high concentration with such monomers, followed by exposure to a higher wavelength UV light (UV365), cleaved the conditional ligand and replaced it with the added immunogenic peptides (Rodenko et al., 2006). The exchange by different peptides was performed in separate identifiable wells of a microtiter plate, to unambiguously assess their MHC stabilization.
The described protocol can be employed to identify immunogenic epitopes of diverse antigens of a pathogen, cancers, as well as autoantigens; but the availability of cells deficient in antigen-processing machinery for the concerned species, as well as the generation of recombinant proteins for HC of class I MHC, and β2m would be critical. Furthermore, the final validation has to be performed by enumerating antigen-specific CD8+ T cells in the host. Therefore, a combinatorial approach could yield validated information on the immunogenicity of epitopes. Another limitation of such protocols might be the inherent polymorphism in MHC molecules occuring in an outbred population. Nonetheless, essential features of T cell differentiation are better captured if the information on immunogenic epitopes of an antigen is available. Further characterization of differentiating antigen-specific CD8+ T cells adds value to such analyses.
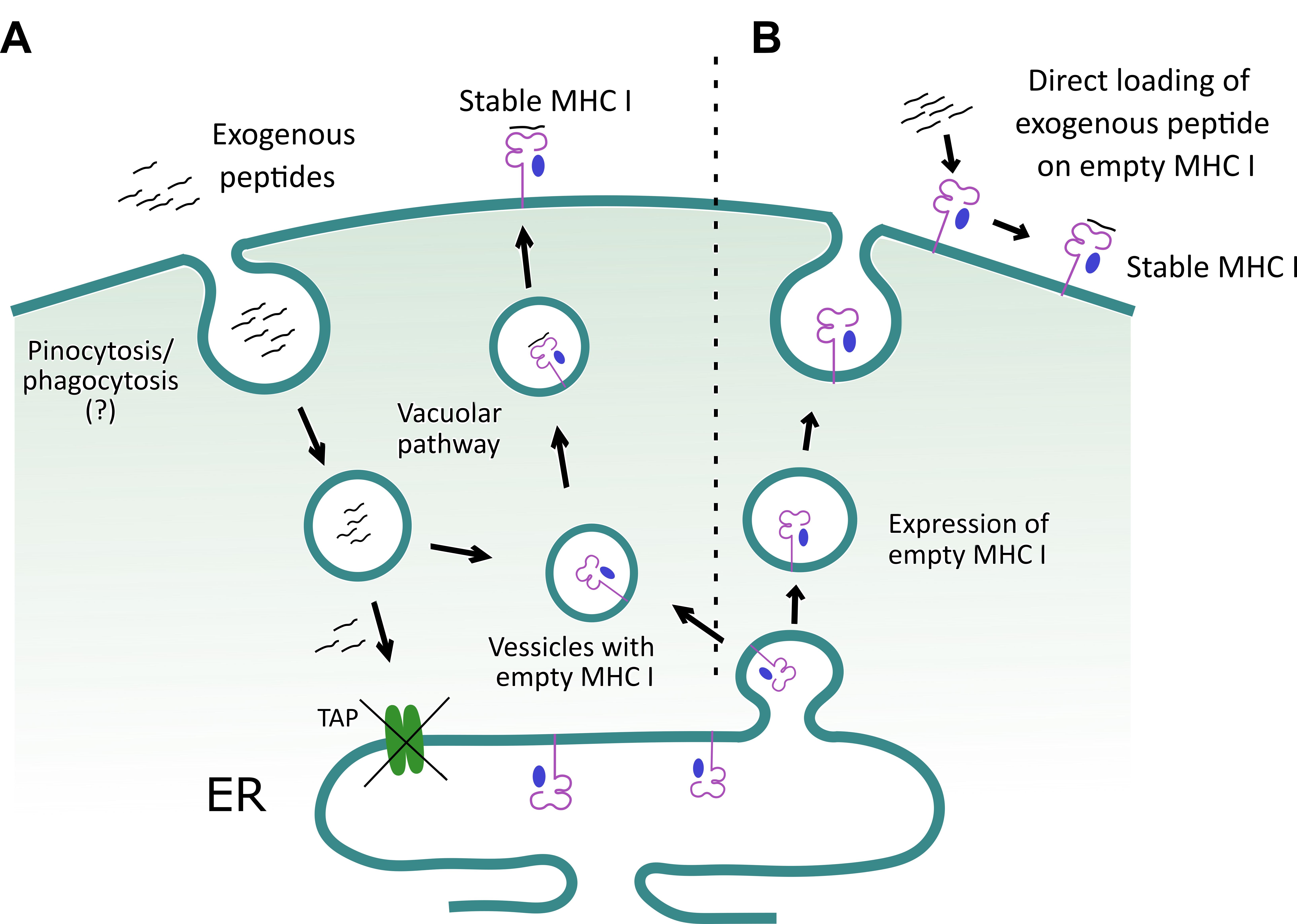
Figure 2. A schematic to depict how exogenous peptides help stabilize class I MHC molecule on the surface of TAP deficient RMA/S cells. A. Exogenous peptides are internalized and loaded onto class I MHC molecules in a yet to be clearly defined subcellular compartment. Subsequently, the loaded class I MHC complexes are exported to the cell surface. The surface stabilized class I MHC molecules are then detected by flow cytometry, using an anti-class I MHC (H-2Kb) antibody. B. Alternatively, RMA/s cells can display empty class I MHC molecule on their surface, and their loading with the exogenously added peptides helps to stabilize expression.
Materials and Reagents
1,000 µl tips (Tarsons, catalog number: 521020)
Racked filter tips, 1,000 µl (Tarsons, catalog number: 528104)
200 µl tips (Tarsons, catalog number: 521014)
Racked filter tips, 200 µl (Tarsons, catalog number: 528106)
10 µl tips (Tarsons, catalog number: 521000)
Racked filter tips, 10 µl (Tarsons, catalog number: 528100)
0.22 µm filtration units (Corning, catalog number: 431118)
5 L flasks (Borosil)
50 ml Centrifuge tubes (Tarsons, catalog number: 500041)
1.5 ml microcentrifuge tubes (Tarsons, catalog number: 500010)
Flat-bottom 96 well cell culture plate (Corning, catalog number: CLS3599-100EA)
Amicon concentrators (Merck, catalog number: UFC903024)
RMA/s cells (NCCS, Pune)
6-8 weeks old male and female C57BL/6 mice (Jackson Laboratories, catalog number: 000664)
H-2Kb and β2m plasmid constructs (Xu et al., 2016) (A kind gift from Hidde L. Ploegh Lab, Boston Children’s Hospital, Harvard University)
Peptides (GL Biochem, Shanghai)
Dimethyl sulfoxide, DMSO (Serva, catalog number: 20385.01)
D-(+)-Glucose anhydrous, Hi-ARTM/ACS (Himedia, catalog number: GRM077-500G)
Ampicillin sodium salt (Himedia, catalog number: MB104-5G)
Isopropyl β-d-1-thiogalactopyranoside, IPTG (BR Biochem Life sciences, catalog number: BC0168-100G)
Lysozyme (Himedia, catalog number: MB098-5G)
DNase I (Sigma-Aldrich, catalog number: 11284932001)
Magnesium Chloride, MgCl2 (Merck, catalog number: 105833)
Ethylene diamine tetraacetic acid, EDTA (Merck, catalog number: 324503)
Sodium-Azide (Merck, catalog number: 8223350100)
Tris-Hydrochloride, Tris-HCl (Himedia, catalog number: GRM1218-5KG)
Sucrose (Duchefa Biochemie, catalog number: S0809.1000)
Diothiothriotol, DTT (Himedia, catalog number: RM525-5G)
Triton X-100 (Serva, catalog number: 37240.01)
Tween 20 (Himedia, catalog number: MB067-100ML)
Sodium Chloride, NaCl (Himedia, catalog number: MB023-500G)
Sodium Hydroxide, NaOH (Himedia, catalog number: MB095-500G)
Di-sodium hydrogen phosphate, Na2HPO4 (Himedia, catalog number: GRM3960-500G)
Potassium dihydrogen phosphate, KH2PO4 (Himedia, catalog number: GRM1188: 500G)
Potassium Chloride, KCl (Merck, catalog number: 1049330500)
Urea (Himedia, catalog number: MB032-5KG)
L-Arginine (Sigma-Aldrich, catalog number: A5006)
Oxidised Glutathione (Himedia, catalog number: RM550-500MG)
Reduced Glutathione (Himedia, catalog number: RM234-5G)
Guanidine-hydrochloride (Serva, catalog number: 39558.02)
Sodium acetate, C2H3NaO2 (Himedia, catalog number: MB048-500G)
Ammonium Chloride, NH4Cl (Merck, catalog number: 1011450500)
Sodium bicarbonate, NaHCO3 (Serva, catalog number: 30180.02)
Polymethylsulphonyl fluoride, PMSF (Serva, catalog number: 32395.04)
4-Nitrophenyl phosphate disodium salt hexahydrate (Sigma-Aldrich, catalog number: 71768-25G)
Propidium iodide, PI (BD biosciences, catalog number: 556463)
Luria Bertani broth, LB broth, Miller (Himedia, catalog number: M1245-1KG)
LB media broth components include:
Tryptone
Yeast extract
Sodium Chloride
BirA biotin-protein ligase standard reaction kit (Avidity)
Kit components include:
d-Biotin (500 µM)
Biotin ligase BirA enzyme (1 mg/ml)
Solution A (bicine buffer: 0.5 M bicine, pH 8.3)
Solution B (ATP biotin: 100 mM ATP, 100 mM MgOAc, 200 µM biotin)
Purified Streptavidin (ThermoFisher, catalog number: SNN1001)
APC conjugated Streptavidin (Biolegend, catalog number: 405207)
RPMI (Gibco, catalog number: 31800-022)
Fetal bovine serum, FBS (Gibco, catalog number: 26140-079)
Penicillin-Streptomycin (Gibco, catalog number: 10378-016)
Anti-mouse FITC-H-2Kb antibody (BD Pharmingen, Clone: AF6-88.5, catalog number: 562002)
Anti-Mouse IgG (whole molecule)-Alkaline phosphatase antibody produced in goat (Sigma-Aldrich, catalog number: A9316-1ML)
Anti-mouse CD8 antibody (clone 53-6.7, BD Bioscience)
Freund’s Adjuvant, complete (Sigma-Aldrich, catalog number: F5881-10ml)
Freund’s Adjuvant, incomplete (Sigma-Aldrich, catalog number: F5506-10ml)
PPRV vaccine strain Sungri/96
Stock solutions (see Recipes)
LB media broth (see Recipes)
Phosphate buffered saline, 1× PBS (see Recipes)
PBS-T (see Recipes)
FACS Buffer (see Recipes)
Resuspension buffer (see Recipes)
Lysis buffer (see Recipes)
Washing buffer with Triton (see Recipes)
Washing buffer without Triton (see Recipes)
Urea buffer (see Recipes)
Refolding buffer (see Recipes)
Guanidine solution (see Recipes)
Gel filtration buffer (see Recipes)
Red blood cell (RBC) Lysis Buffer (see Recipes)
Equipment
Serological and micropipettes (Eppendorf Research Plus)
Spectrophotometer (GeneQant, model: 80-2130-00)
Autoclave (ALP, model: CL-40MDP)
Incubator with shaker (TBOY 5000I INCUB SHKER, catalog number: 980473)
Sonicator (Sonics & Materials INC., model: VCX750)
Centrifuges (Eppendorf, models: AG 5810 R and 5401)
Gel filtration apparatus (AKTA) with cold chamber (HiPrep 16/60, Sephacryl S200 HR)
UV-chamber (Genetix, model: GX-2082-6111)
BD Accuri C6 Flow cytometer (BD Biosciences)
SDS gel apparatus (Bio Rad Mini-PROTEAN Tetra System)
Eon ELISA Plate Reader (BioTek Instruments, Inc.)
LSE Vortex Mixer (Corning, catalog number: 6776)
Laboratory Stirrer/Hot Plate (Corning, catalog number: 6798-420D)
Magnetic bars (Tarsons, catalog number: 4117)
Software
UCSF Chimera version 1.13 (https://www.cgl.ucsf.edu/chimera) (Pettersen et al., 2004; Chen et al., 2015)
HPEPDOCK online docking tool (http://huanglab.phys.hust.edu.cn/hpepdock/) (Zhou et al., 2018)
NCBI, PDB, Uniprot protein Databases (https://www.ncbi.nlm.nih.gov/; https://www.rcsb.org/; https://www.uniprot.org/)
Immune Epitope Database and Analysis Resource (IDEB) and IDEB-AR (Zhang, 2013)
FlowJo vX (Becton Dickinson and company)
GraphPad Prism v8.4.3 (San Diego, CA)
Microsoft office 2016
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sarkar, R., Sharma, Y., Jain, A., Tehseen, A., Singh, S. and Sehrawat, S. (2021). A Combinatorial in-silico, in-vitro and in-vivo Approach to Quantitatively Study Peptide Induced MHC Stability. Bio-protocol 11(24): e4255. DOI: 10.21769/BioProtoc.4255.
分类
免疫学 > 免疫细胞染色 > 流式细胞术
免疫学 > 动物模型 > 小鼠
生物科学 > 生物技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













