- EN - English
- CN - 中文
Isolation of Plant Nuclei Compatible with Microfluidic Single-nucleus ATAC-sequencing
与微流体单核 ATAC 测序兼容的植物核的分离
发布: 2021年12月05日第11卷第23期 DOI: 10.21769/BioProtoc.4240 浏览次数: 4994
评审: Wenrong HeYuan WangTravis Lee
Abstract
Gene expression depends on the binding of transcription factors with DNA regulatory sequences. The level of accessibility for these sequences varies between cells and cell types. Until recently, using the Tn5 assay for transposase-accessible chromatin for sequencing (ATAC-seq) technology allowed assessing the profiles of chromatin from an entire organ or, when coupled with the isolation of nuclei tagged in specific cell types (INTACT) method, from a cell-type. Recently, ATAC-seq experiments were conducted at the level of individual plant nuclei. Applying single nuclei ATAC-seq (sNucATAC-seq) technology to thousands of individual cells revealed more finely tuned profiles of chromatin accessibility. In this manuscript, we describe a method to isolate nuclei fom plant roots and green tissues, permeabilize the nuclear membrane using detergent to allow the penetration of the Tn5 transposase, and re-suspend them in a nuclei resuspension buffer compatible with the construction of sNucATAC-seq libraries using the 10× Genomic’s Chromium technology. This protocol was successfully applied on Arabidopsis thaliana and Glycine max root nuclei.
Background
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) is a popular technology to map DNA accessible sites and to identify putative regulatory elements (Lu et al., 2017; Huang et al., 2021). The accessibility of these elements plays an important role in controlling gene expression because they are transcription factor-binding sites. One major limitation associated with the use of ATAC-seq technology is the cellular complexity of the sample used. When working at the entire organ level, the profile of chromatin accessibility cannot reveal the differential and unique contributions of each cell and cell type composing the organ. To overcome this challenge, recent studies successfully tested ATAC-seq technology on thousands of individual plant nuclei isolated from Arabidopsis thaliana roots and various Zea mays organs (Dorrity et al., 2021; Farmer et al., 2021; Marand et al., 2021).
The integration of this molecular information with RNA-seq datasets generated from isolated protoplasts and nuclei revealed the impact of chromatin accessibility and regulatory elements in controlling gene expression (Denyer et al., 2019; Jean-Baptiste et al., 2019; Nelms and Walbot, 2019; Ryu et al., 2019; Shulse et al., 2019; Zhang et al., 2019; Dorrity et al., 2021; Farmer et al., 2021; Liu et al., 2020; Marand et al., 2021). Therefore, single-cell/nucleus RNA-seq and single nuclei ATAC-seq (sNucATAC-seq or snATAC-seq) have recently changed our assessment of the molecular mechanisms controlling gene expression. These technologies will also contribute to a better understanding of gene function to control plant cell differentiation, development, and plant cell response to environmental stresses (Rich-Griffin et al., 2020; Shaw et al., 2021).
The SNucATAC-seq technology relies on the permeabilization of the membrane of isolated plant nuclei to enable the penetration of the Tn5 transposase, the tagmentation of the accessible genomic DNA sequences, and the construction of Illumina-compatible libraries. In this manuscript, we describe the methodology used to conduct sNucATAC-seq. This method complements the protocol describing the implementation of single nuclei RNA-seq technology on plant samples (Thibivilliers et al., 2020). Here, we describe a protocol to isolate and purify nuclei from model plants and crop species before permeabilizing the nuclear membrane to enable the penetration of the Tn5 transposase (Figure 1). Examples of successful sNucATAC-seq profiles from Arabidopsis thaliana and Glycine max are also provided.
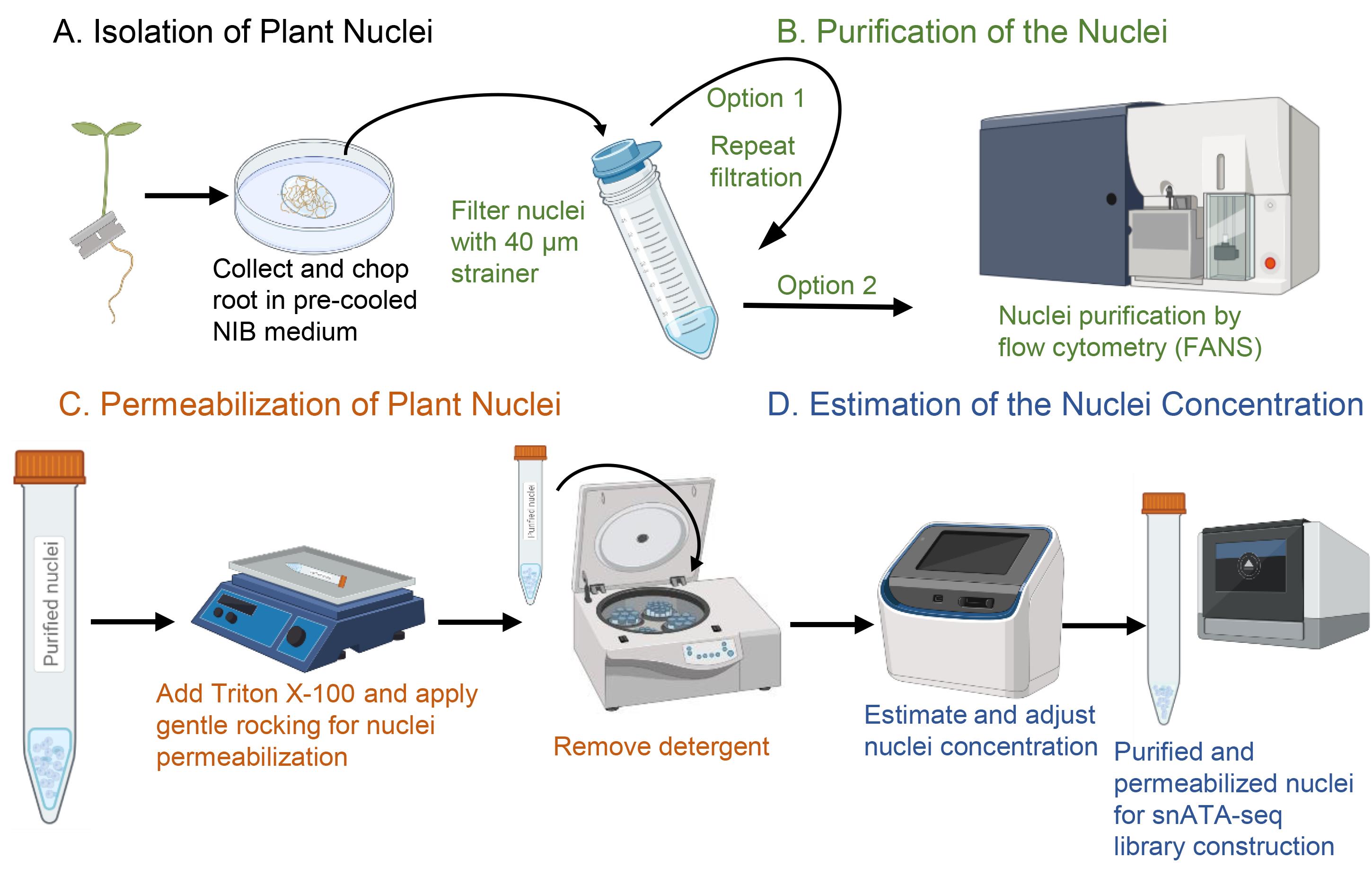
Figure 1. Workflow of nuclei isolation, purification, and permeabilization compatible with single-nucleus ATAC-seq library construction (created in BioRender.com).
Materials and Reagents
60 mm Petri dishes (Corning Life Sciences, catalog number: 08757100B)
Single edge extra keen razor blade (Electron Microscopy Sciences, catalog number: 71962)
50 ml Falcon conical tubes (Corning, catalog number: 352070)
15 ml Falcon conical tubes (Corning, catalog number: 430790)
MACS SmartStrainers 30 µm (Miltenyi Biotec, catalog number: 130-098-458)
40 µm sterile cell strainers (Corning, catalog number: 07-201-430)
Mini 20 µm cell strainers (pluriStrainer, catalog number: 43-10020-50)
Micro slide (Corning, catalog number: 2940-75x25)
Cover glass (Thermo Scientific, catalog number: 12450S)
Eppendorf microcentrifuge tube (Fisher Scientific, catalog number: 05-402-25)
Pipette tips [Rainin RT LTS, catalog numbers: 30389226 (10 µl), 30389240 (200 µl), 30389213 (1,000 µl), 30389218 (1,000 µl wide-orifice)]
CelLyticTM PN Isolation/Extraction buffer 4× (Sigma-Alrich, catalog number: CELLYTPN1) is diluted with DNAse free water to reach a final working concentration of 1×. We name this diluted buffer “NIB medium.”
Nuclease-free water (Invitrogen, catalog number: AM9937)
Phosphate-buffered saline (PBS) without calcium and magnesium (Corning, catalog number: 21-040-CV)
Bovine serum albumin 50 mg/ml (BSA; Invitrogen, catalog number: AM2616)
7-AAD (eBioscience, catalog number: 00-6993-50)
SYTOX Green nucleic acid stain (Invitrogen, catalog number: S7020)
Triton X-100 (Sigma, catalog number: T8787-100ml)
Nuclei buffer 20× (10× Genomics, catalog number: 2000207; part of the Chromium Next GEM Single Cell ATAC Library Kit v1.1, PN-1000163/4)
Trypan blue stain 0.4% (Invitrogen, catalog number: T10282)
1% BSA-PBS (see Recipes)
NIB medium supplemented with 2% Triton X-100 (see Recipes)
1% BSA-PBS supplemented with 2% Triton X-100 (see Recipes)
Equipment
Dissecting Forceps, Fine Tip (VWR, catalog number: 82027-386)
Countess cell counting chamber slide (Invitrogen, catalog number: C10228)
BD FACSAria II cell sorter (BD Biosciences, San Jose, CA)
Microvolume pipettes (Rainin, models: Lite – XLS, P1000, P200, P20)
Platform Rocker (Fisherbrand, catalog number: 13-687-704)
Sorvall Legend X1R Centrifuge with Swinging Bucket Rotor (Thermo Scientific, catalog number: 75004261)
Countess II Automated Cell Counter (Invitrogen, catalog number: AMQAX1000)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Thibivilliers, S. B., Anderson, D. K. and Libault, M. Y. (2021). Isolation of Plant Nuclei Compatible with Microfluidic Single-nucleus ATAC-sequencing. Bio-protocol 11(23): e4240. DOI: 10.21769/BioProtoc.4240.
分类
植物科学 > 植物分子生物学 > DNA > DNA 结构
分子生物学 > DNA > 染色质可接近性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link