- EN - English
- CN - 中文
Live Imaging of Apoptotic Extrusion and Quantification of Apical Extrusion in Epithelial Cells
上皮细胞凋亡挤压的活体成像和根尖挤压的定量分析
发布: 2021年11月20日第11卷第22期 DOI: 10.21769/BioProtoc.4232 浏览次数: 3758
评审: Andrea PuharJan HuebingerDana Manuela Savulescu
Abstract
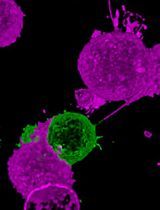
Apoptotic cell death eliminates unhealthy cells and maintains homeostatic cell numbers within tissues. Epithelia, which serve as fundamental tissue barriers for the body, depend on a physical expulsion of dying cells (apoptotic cell extrusion) to remain sealed and intact. Apoptotic cell extrusion has been widely studied over recent years, with researchers using various approaches to induce apoptotic cell death. Unfortunately, the majority of chemical-based approaches for cell death induction rely on sporadically occurring apoptosis and extrusion, making imagining lengthy, often unsuccessful, and difficult to capture in high-quality images because of the frequent frame sampling needed to visualise the key molecular processes that drive extrusion. Here, we present a protocol that describes steps needed for laser-mediated induction of apoptosis in a cell of choice, followed by imaging of apoptotic extrusion in confluent monolayers of epithelial cells. Moreover, we provide the description of a new approach involving the mixing of labelled and unlabelled cells. In particular, this protocol characterises how cells surrounding apoptotic cells behave, with high spatial and temporal resolution. This can be achieved without the optical interference that apoptotic cells cause as they are physically expelled from the monolayer and move out of focus for imaging. Finally, the protocol is accompanied by detailed procedures describing cell preparation for apoptotic extrusion experiments, as well as post-acquisition analysis required to evaluate rates of successful extrusion.
Keywords: Epithelia (上皮细胞)Background
Epithelial tissues rely on effective extrusion to eliminate apoptotic cells for the preservation of tissue barriers and to prevent inflammation. As homeostatic apoptosis is a relatively sporadic event in healthy cultured epithelia, apoptosis needs to be experimentally induced to reliably visualise the extrusion phenomenon. This may be achieved by a variety of methods, including exposure to UV radiation (Rosenblatt et al., 2001; Slattum et al., 2009; Andrade and Rosenblatt, 2011), treatment with various chemicals [e.g., Etoposide (Andrade and Rosenblatt, 2011; Michael et al., 2016; Teo et al., 2020), Camptothecin (Andrade and Rosenblatt, 2011), or activation of Fas receptors (Andrade and Rosenblatt, 2011)]. However, most commonly used approaches rely on randomly-induced apoptosis, making it challenging to capture high quality images with high temporal resolution to characterise dynamically progressing extrusion events. The protocol presented here describes induction of apoptosis in pre-selected cells of choice, thus allowing continuous high-quality imaging of apoptotic extrusion from induction of cell death to completion of extrusion. The following protocol may be adopted to selectively induce apoptosis in other models [e.g., Zebrafish periderm (Duszyc et al., 2021)] and can also be used for detailed analysis of the behaviour of neighbouring cells during the extrusion process by selectively labelling neighbours with genetically encoded fluorescent proteins (Duszyc et al., 2021).
Materials and Reagents
40 µm cell strainers (Biologix, catalog number: 15-1040)
Glass bottom dishes, No. 1.5 Coverslip (35 mm diameter, MATTEK, catalog number: P35G-1.5-20-C) or an equivalent (e.g., ibidi, catalog number: 81158)
75 cm2 tissue culture treated flasks with filter caps (Thermo Fisher Scientific, NuncTM EasYFlaskTM catalog number: NUN156499) or an equivalent (e.g., Corning, catalog number: CLS430641U)
Disposable serological pipettes (Corning® Costar® Stripette®, catalog numbers: CLS4487 [5 ml]; CLS4488 [10 ml])
Sterile filter tips (ZAPTM, catalog numbers: 1051-965-038 [0.1-10 µl]; 1055-965-018 [1-40 µl]; 1059-965-008 [1-200 µl]; 1057-965-018 [1,000 µl])
1.5 ml Eppendorf tubes (Eppendorf South Pacific Pty Ltd, catalog number: 0030120086)
Centrifuge Falcon Tubes (BD FalconTM, catalog numbers: BDAA352096 [15 ml], BDAA352070 [50 ml])
Epithelial cell lines with well-defined cell-cell junctions such as MCF-7 or Caco-2: wild type and expressing fluorescently tagged proteins that can mark cell-cell junctions (e.g., E-cadherin or ZO-1), the cytoskeletal cortex at cell-cell junctions (e.g., Myosin regulatory light chain, MRLC) or biosensors of junctional proteins (e.g., active RhoA biosensor AHPH). Green (e.g., GFP) or red (e.g., mCherry) fluorescent proteins should be used as fluorescent tags to distinguish from far-red Annexin V staining (Alexa FluorTM 647)
Fetal bovine serum (FBS) (HyCloneTM, catalog number: SH30084.03)
Annexin V, Alexa FluorTM 647 conjugate (Thermo Fisher Scientific, catalog number: A23204)
Opti-MEM media (Thermo Fisher Scientific, catalog number: 31985070)
LipofectamineTM RNAiMAX transfection Reagent (Thermo Fisher Scientific, catalog number: 13778150)
MEM Non-Essential Amino Acids 100× (MEM NEAA; Thermo Fisher Scientific, GibcoTM, catalog number: 11140050)
L-glutamine 100× (Thermo Fisher Scientific, GibcoTM, catalog number: 25030149)
Penicillin-Streptomycin (5,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15070063)
DMEM for MCF-7 cells (Thermo Fisher Scientific, GibcoTM, catalog number: 11995073) or RPMI for Caco-2 cells (Thermo Fisher Scientific, GibcoTM, catalog number: 21870076)
Hank’s balanced salt solution (Sigma-Aldrich, catalog number: H8264)
Complete cell culture medium (see Recipes)
Imaging medium (see Recipes)
Equipment
Pipet gun (e.g., Thermo Fisher ScientificTM, catalog number: 9501)
Set of pipettes (e.g., Gilson PIPETMAN®, catalog numbers: FA10001M [0.2-2 µl]; FA10003M [2-20 µl]; FA10005M [20-200 µl]; FA10006M [100-1,000 µl])
Bright field microscope (e.g., Nikon Eclipse TS100 equipped with Plan Fluor 4×/0.13 phase contrast objective)
Hemocytometer counting chamber (e.g., Livingstone, catalog number: HCCB)
Cell culture CO2 incubator (e.g., PHCbi catalog number: MCO-230AICUVL-PA)
Class II biological safety cabinet (e.g., Gel Aire, model: 20 BHEN 2004D)
Benchtop centrifuge (e.g., Hettich Lab TechnologyTM ROTIFIX 32A)
Laser scanning confocal microscope minimally equipped with:
Dichroic and emission filters for the use of the 488 nm and 633nm laser lines and detection of GFP and Alexa FluorTM 647 fluorescence, respectively
488nm and 633 nm laser lines (e.g., Argon, HeNe or solid-state lasers)
Acoustic-optic tunable filters (AOTF) for irradiation of a specific region of interest (i.e., bleaching of selected areas)
2W or more tunable two-photon laser (adjustable to 790nm laser line) (e.g., MaiTai or Chameleon)
Heated chamber (37°C) for live cell imaging
40× IR-VIS compatible objective (ideally water immersion to minimise chromatic aberrations)
The experiments shown in this protocol were performed on a Zeiss LSM 710 Meta Confocal Scanner equipped with a fully tunable Mai Tai eHP DeepSee 760-1040 nm laser, heated chamber (37°C for cell imaging), and Plan Apochromat 40× 1.3 N.A. water objective.
Software
ImageJ software (https://imagej.nih.gov/ij/)
GraphPad PRISM software (http://www.graphpad.com)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Duszyc, K., Noordstra, I., Yap, A. S. and Gomez, G. A. (2021). Live Imaging of Apoptotic Extrusion and Quantification of Apical Extrusion in Epithelial Cells. Bio-protocol 11(22): e4232. DOI: 10.21769/BioProtoc.4232.
分类
细胞生物学 > 细胞成像 > 活细胞成像
细胞生物学 > 细胞活力 > 细胞死亡
发育生物学 > 细胞生长和命运决定
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link