- EN - English
- CN - 中文
Assessing the in vitro Binding Specificity of Histone Modification Reader Proteins Using Histone Peptide Arrays
利用组蛋白肽阵列评估组蛋白修饰阅读蛋白的体外结合特异性
发布: 2021年09月20日第11卷第18期 DOI: 10.21769/BioProtoc.4168 浏览次数: 3513
评审: Laxmi Narayan MishraYalong Aaron WangAnonymous reviewer(s)

相关实验方案

基于蛋白筛选策略从合成文库中分离抗原特异性纳米抗体:结合 MACS 的酵母展示筛选与 FLI-TRAP 方法
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
2026年01月20日 475 阅读

用于筛选 SLIT2 结合分子的正交温度相关荧光强度变化与时间分辨高通量筛选平台
Moustafa T. Gabr [...] Somaya A. Abdel-Rahman
2026年02月20日 104 阅读
Abstract
In the field of chromatin biology, a major goal of understanding the roles of histone post-translational modifications is to identify the proteins and domains that recognize these modifications. Synthetic histone peptides containing one or more modifications are a key tool to probe these interactions in pull-down assays with recombinant proteins or cell lysates. Building on these approaches, the binding specificity of a protein of interest can be screened against many histone peptides in parallel using a peptide array. In this protocol, we describe the expression and purification of a recombinant protein of interest in bacteria, followed by an assay for binding to histone post-translational modifications using a commercially available histone peptide array. The purification uses a versatile dual-tagging and cleavage strategy and equipment commonly available in a molecular biology laboratory.
Graphic abstract:
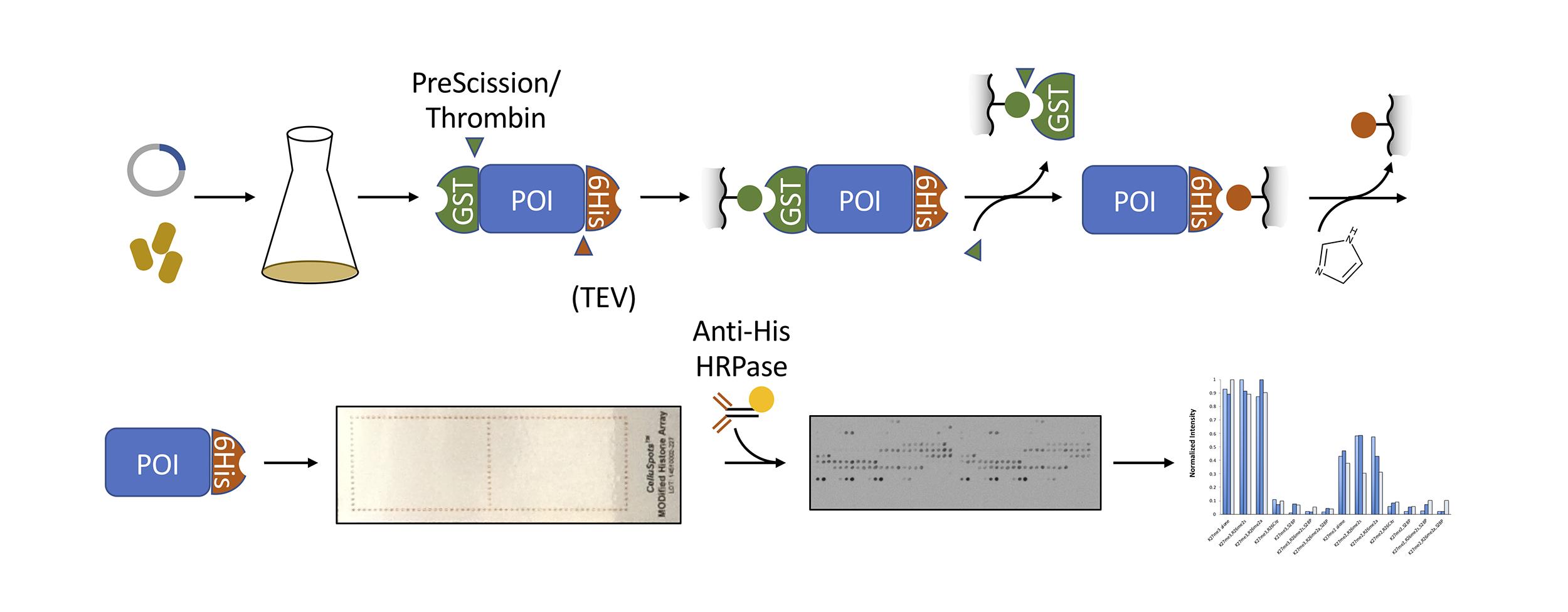
Overview of protocol for purifying recombinant protein and hybridizing to a histone peptide array.
Background
The histone proteins that package the eukaryotic genome can exhibit covalent post-translational modifications, including methylation, phosphorylation, and acetylation, at numerous residues. The functions of some of these modifications in chromatin regulation are mediated by interactions with effector proteins containing so-called ‘reader’ domains. To date, many reader domains have been characterized, including the CHROMO (chromatin organization modifier), Tudor, PWWP, MBT (Malignant Brain Tumour) repeat, PHD (plant homeodomain), ankyrin repeat, ADD (ATRX-DNMT3-DNMT3L), YEATS (Yaf9, ENL, AF9, Taf14, Sas5), WD40, and bromodomains (reviewed in Musselman et al., 2012). These domains are present in diverse proteins with roles in fundamental processes, including gene regulation, DNA repair, recombination, and replication. However, the full catalogue of chromatin reader proteins remains to be uncovered.
Early biochemical and structural studies of histone modification reader proteins revealed modification-dependent interactions with short histone peptides (Dhalluin et al., 1999; Jacobson et al., 2000; Bannister et al., 2001; Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). In addition, many of the characterized histone modifications occur on the N- or C-terminal histone ‘tail’ domains, which are intrinsically disordered and likely to be conformationally dynamic in the absence of interaction partners (reviewed in Ghoneim et al., 2021). Thus, when assaying interactions of reader proteins with histone modifications, a short histone peptide with the modification and its neighbouring sequence is likely to be an effective proxy for the native histone protein.
The introduction of peptide arrays to the chromatin field provided a powerful method to assay the binding of a protein of interest to many different modifications simultaneously in parallel (Nady et al., 2008). Since then, several types of commercially available histone peptide arrays have been developed, and their characteristics have been thoroughly reviewed recently (Mauser and Jeltsch, 2019). In this protocol, we use the MODifiedTM histone peptide array (Active Motif) (Bock et al., 2011), which contains 384 unique peptide spots in duplicate, synthesized using the CelluSpots method (Winkler et al., 2009). The cellulose-bound 19mer peptides on the array represent sequences from different regions of the core histones (H3 amino acids # 1-19, 7-26, 16-35, 26-45, H4 1-19, 11-30, and H2A, H2B 1-19) as well as five control spots. The peptides are either unmodified, contain a single modification, or a combination of up to 4 modifications on the same peptide. A total of 59 modifications are represented, including acetylation, mono-, di- and tri-methylation of lysine, symmetric and asymmetric dimethylation of arginine, phosphorylation, and citrullination of different residues. A limitation of the array approach is that interaction detection is restricted to the particular modifications, combinations, and peptide sequences present on the array.
Heterologous protein expression in E. coli and affinity purification is a widely used strategy to obtain protein for in vitro assays. The protocol described here is adapted from standard protocols (for a detailed discussion, see Harper and Speicher, 2011). We designed plasmids for tandem purification with protease-cleavable tags at both the N- and C-termini. The N-terminal glutathione S-transferase (GST) fusion aids in solubilization and is used for the first affinity purification on glutathione-sepharose resin. The GST is then cleaved by PreScission Protease (or thrombin) to elute the recombinant protein, which contains a C-terminal 6×Histidine (His) tag. The 6×His tag is used for a second purification on an immobilized metal resin, as well as for antibody detection on the histone peptide array. We have used this versatile 2-step purification strategy and histone peptide array protocol to examine the binding specificities of several C. elegans chromo domain-containing proteins (Saltzman et al., 2018).
Materials and Reagents
Note: Equivalent materials and reagents can be used.
Clone cDNA into GST/His expression vector (pALS099 or pALS343)
Chemically competent E. coli cloning strain (e.g., DH5α, Invitrogen, catalog number: 18265017 or equivalent)
High-fidelity DNA polymerase (e.g., Q5, New England Biolabs, catalog number: M0491S; Phusion, New England Biolabs, catalog number: M0530S)
Gel-purification kit (e.g., GeneAid, catalog number: DFH300)
Template for cDNA of interest or a cDNA clone
Reagents for cloning method of choice:
(Option 1) Reagents for Gibson assembly cloning:
Plasmid pALS343 (pGEX-2TP-cTevHis-Gibson/MCS; Addgene, catalog number: 139788)
FastDigest PdiI restriction enzyme (Fisher Scientific, catalog number: FD1524)
FastDigest KpnI restriction enzyme (Fisher Scientific, catalog number: FD0524)
NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, catalog number: E2621) or homemade Gibson assembly master mix (Gibson et al., 2009)
(Option 2) Reagents for GatewayTM cloning
Plasmid pALS099 (pGEX-2TP-cTevHis-DEST; Addgene, catalog number: 139787)
A GatewayTM donor vector (non-ampicillin resistant; e.g., pDONR221, Fisher Scientific, catalog number: 12536017)
LR ClonaseTM II enzyme mix (Invitrogen, catalog number: 11791-020)
Express tagged protein
Nalgene PPCO centrifuge bottles with sealing closure (500 ml; Fisher Scientific, catalog number: 3141-0500 or equivalent, matched with available rotor – see Equipment)
0.22 μm filter bottles, 500 ml (Fisher Scientific, catalog number: 09-741-05)
0.22 μm filter units, 50 ml (Millipore Sigma, catalog number: SCGP00525)
Expression plasmid(s) cloned above
Chemically competent E. coli protein expression strain, e.g., BL21(DE3)pLysS (Invitrogen LSC606010) or C41(DE3)pLysS (Novagen 60444-1) (see Notes)
LB-Agar plates containing 100 μg/ml ampicillin and additional antibiotics as required for your expression strain
Dehydrated 2× YT media (Fisher Scientific, BD DifcoTM, catalog number: DF0440-17-0)
Glucose (Sigma-Aldrich, catalog number: G7021)
Isopropyl-β-D-thiogalactoside (IPTG, MW 238.3 g/mol) (Calbiochem, catalog number: 420322)
20% (w/v) Glucose (see Recipes)
2× YTG Media (see Recipes)
0.5 M IPTG (See Recipes)
Purify tagged protein
Cell pellet from above
Nalgene Oak Ridge Style Round Bottom Centrifuge Tube (Fisher Scientific, catalog number: 3114-0050 or equivalent, matched to available rotor – see Equipment)
Empty small chromatography columns (e.g., Bio-Rad Biospin chromatography columns, catalog number: 732-6008)
Glutathione Sepharose 4B (Cytiva – formerly GE Life Sciences, catalog number: 17075601) (store at 4°C)
GST-tagged PreScission Protease (Cytiva – formerly GE Life Sciences, catalog number: 27084301) (store at -20°C)
TALON Metal Affinity Resin (Cobalt Sepharose 6B-CL, Takara, catalog number: 635501) (store at 4°C)
Fast Macro DispoDialyzer, cutoff > 10kDa (Harvard Apparatus, catalog number: 74-0802)
NaCl (Sigma-Aldrich, catalog number: S3014)
KCl (Sigma-Aldrich, catalog number: P9541)
Tween-20 (Sigma-Aldrich, catalog number: P9416)
Triton X-100 (Sigma-Aldrich, catalog number: T8787)
Phenylmethanesulfonyl fluoride (PMSF) (Roche, catalog number: 10837091001)
cOmpleteTM mini EDTA-free Protease inhibitor cocktail tablets (Roche, catalog number: 4693159001) (store at 4°C)
NaH2PO4 (119.98 g/mol) (Sigma-Aldrich, catalog number: S0751)
Na2HPO4 (141.96 g/mol) (Sigma-Aldrich, catalog number: S7907)
Imidazole (68.08 g/mol) (Sigma-Aldrich, catalog number: I5513)
37% Hydrochloric Acid (HCl) (Sigma-Aldrich, catalog number: 258148)
1 M HEPES-NaOH pH 7.5 (Tekno, catalog number: H1035)
0.5 M EDTA pH 8.0 (Invitrogen, catalog number: LS15575020)
2-mercaptoethanol (2-ME) (14.3 M) (Sigma-Aldrich, catalog number: M3148) (store at 4°C)
1 M DTT solution (Sigma-Aldrich, catalog number: 43816) (aliquot and store at -20°C)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Reagents for SDS-PAGE (homemade gels or other suppliers can be substituted):
4× Laemmli protein sample buffer (Bio-Rad, catalog number: 161-0747)
Precast Protein Gels (e.g., Bio-Rad Mini-PROTEAN® TGXTM of appropriate percentage for your protein)
10× Tris-Glycine-SDS running buffer (Bio-Rad, catalog number: 161-0732)
Precision Plus Protein Dual Xtra Prestained Protein Standards (Bio-Rad, catalog number: 161-0377)
Coomassie Brilliant Blue R-250 (Fisher Scientific, catalog number: BP101)
Protein quantification reagent (e.g., Bradford assay; Bio-Rad, catalog number: 500-0006)
Stock Solutions (see Recipes)
5 M NaCl
2 M KCl
10% (v/v) Triton X-100
10% (v/v) Tween-20
100 mM PMSF
1 M NaH2PO4
0.5 M Na2HPO4
100 mM Sodium Phosphate buffer, pH 8.0
(5× Reduced Glutathione Buffer – optional – only required if eluting from the GSH-sepharose rather than cleaving off the GST)
3 M Imidazole pH ~7.5
5× Lysis Buffer
Buffers (see Recipes)
1× Lysis Buffer
GST High Salt Wash Buffer
GST Low Salt Wash Buffer
PreScission Protease Buffer
Talon binding/wash buffer
Talon Elution Buffer
Dialysis Buffer
(Tris pre-elution buffer – optional – only required if eluting from the GSH-sepharose rather than cleaving off the GST)
(Glutathione elution buffer – optional – only required if eluting protein from the GSH-sepharose rather than cleaving off the GST)
Coomassie Staining Solution
Coomassie Destain Solution
Histone Peptide Array assay
Plastic food wrap
Autoradiography ruler (Diversified Biotech ARS-150, Sigma-Aldrich, catalog number: R8133)
Film (Carestream® BioMax® XAR Film, catalog number: 1651454; Sigma-Aldrich, catalog number: F5513)
Film cassette (e.g., Cytiva – formerly GE Life Sciences, catalog number: GERPN11649)
MODifiedTM histone peptide array (Active Motif, catalog number: 13001) (store at -20°C)
Container to incubate the 1 inch × 3 inch array slide (e.g., small plastic boxes used for blot strips – Genhunter B123, Genhunter B130, Nunc 267061, or a slide hybridization chamber)
Purified protein of interest (aliquots stored at -80°C)
Anti-His(C-term)-HRP (Invitrogen, catalog number: R931-25) (store at 4°C)
10× phosphate-buffered saline (PBS) (Fisher Scientific, catalog number: BP399-1)
Skim milk powder (EMD Millipore, catalog number: 115363)
Bovine serum albumin (BSA; Sigma-Aldrich, catalog number: A7906)
Enhanced chemiluminescence reagents (ECL) (e.g., Western Lightning Plus-ECL, Perkin-Elmer NEL103E001EA) (store at 4°C) (see Notes)
Buffers (see Recipes)
Wash buffer (PBS-T)
Blocking Buffer
Pre-binding buffer
Peptide Array binding buffer
Equipment
Note: Equivalent equipment can be used.
Sonicator (e.g., Branson Sonifier 250/450, with appropriate tip size) or similar ultrasonic homogenizer, alternatively a French Press for cell lysis
Refrigerated high-speed centrifuge with rotors for 4,000-7,000 × g for 500 ml bottles and 20,000 × g for Oakridge 30-50 ml tubes (e.g., Beckman Avanti-JE with JA-10 and JA-25.50 rotors; or Sorvall RC-6 with SLA-3000 and SS-34 rotors)
Refrigerated low-speed centrifuge (e.g., as above or Beckman Allegra X-14R with swinging bucket rotor SX4750)
Refrigerated microcentrifuge (e.g., Eppendorf 5424R, or microcentrifuge in a cold room)
Shaker incubator (e.g., New BrunswickTM I24, with refrigeration (I24R) if low induction temperatures required – e.g., 16-18°C)
Cold room (4°C) or glass door refrigerator
Platform shaker (e.g., VWR, model: 3500 Standard Shaker, catalog number: 89032-092)
End-over-end tube rotator/rotisserie (e.g., Fisher Scientific, catalog number: 11676341)
Magnetic stir plate (e.g., Fisher Scientific, catalog number: 1152016S)
Vertical electrophoresis system for SDS-PAGE and Western blotting (e.g., Bio-Rad Mini-PROTEAN tetra cell 1658000)
Spectrophotometer (OD600 and OD595 for protein assay)
Film developer and darkroom (or alternate detection method, e.g., a Bio-Rad Chemidoc 12003153)
Scanner (e.g., Epson Perfection V850 Pro)
Software
Array Analyze Software (Active Motif, https://www.activemotif.com/documents/Array_Analyze_Software_v16.zip)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Soo, M. W. and Saltzman, A. L. (2021). Assessing the in vitro Binding Specificity of Histone Modification Reader Proteins Using Histone Peptide Arrays. Bio-protocol 11(18): e4168. DOI: 10.21769/BioProtoc.4168.
分类
系统生物学 > 表观基因组学 > 组蛋白修饰
生物化学 > 蛋白质 > 相互作用 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link