- EN - English
- CN - 中文
In situ Hybridization of miRNAs in Human Embryonic Kidney and Human Pluripotent Stem Cell-derived Kidney Organoids
在人胚肾和人多能干细胞衍生的肾类器官中微小RNA的原位杂交
发布: 2021年09月05日第11卷第17期 DOI: 10.21769/BioProtoc.4150 浏览次数: 3341
评审: Giusy TornilloAnonymous reviewer(s)
Abstract
MicroRNAs are small RNAs that negatively regulate gene expression and play an important role in fine-tuning molecular pathways during development. There is increasing interest in studying their function in the kidney, but the majority of studies to date use kidney cell lines and assess the total amounts of miRNAs of interest either by qPCR or by high-throughput methods such as next generation sequencing. However, this provides little information as to the distribution of the miRNAs in the developing kidney, which is crucial in deciphering their role, especially as there are multiple kidney cell types, each with its own specific transcriptome. Thus, we present a protocol for obtaining spatial information for miRNA expression during kidney development by in situ hybridization (ISH) of anti-miRNA, digoxigenin-labelled (DIG), Locked Nucleic Acid (LNA®) probes on (i) native human embryonic tissue and (ii) human pluripotent stem cell (hPSC)-derived 3D kidney organoids that model kidney development. We found that the method reveals the precise localization of miRNA in specific anatomical structures and/or cell types and confirms their absence from others, thus informing as to their specific role during development.
Keywords: Kidney development (肾脏发育)Background
MicroRNAs (miRNAs) are small (20-25 nucleotides) RNAs that regulate gene expression by binding predominantly to the 3’ UTRs of their target genes’ mRNAs and inhibiting their translation and/or causing their degradation (Bartel, 2018). An increasing number of studies now show that miRNAs play a crucial role in fine-tuning molecular pathways in the kidney, both in normal development and in disease (Jones et al., 2018; Trionfini et al., 2015; Zhao et al., 2019). The majority of these studies have been conducted on animal models, which do not always faithfully recapitulate the developmental events or disease phenotypes in humans. To overcome these limitations, we recently reported using human pluripotent stem cell (hPSC)-derived kidney organoids (Bantounas et al., 2018; Takasato et al., 2015) as a model to study the role of the miR-199a/214 cluster during human kidney development (Bantounas et al., 2021). In that study, we used in situ hybridization of miRNAs with digoxigenin-labelled locked-nucleic acid (LNA®) probes to detect the precise localization of the miRNAs of this cluster in paraffin sections of both native kidney and organoids. LNA® probes have a chemically modified backbone to afford a higher melting temperature (Tm) per nucleotide than conventional DNA or RNA probes (Singh et al., 1998). They are, thus, ideally suited to the detection of smaller RNAs for which conventional probes would have too low a Tm to bind efficiently. After binding to the tissue, the probes are detected by application of an alkaline phosphatase (AP)-linked, anti-digoxigenin antibody, followed by the addition of an AP substrate, which is converted to a coloured product, visible under a light microscope. Using this method, we discovered that miR-199a-3p and miR-214-3p were both present in the kidney stroma and in developing glomeruli but were largely absent from mature glomeruli. One of the miRNAs, miR-214-3p, also exhibited strong tubular expression. In addition, we observed differences in the extent and distribution of expression between embryonic/fetal kidney and organoids (particularly in the case of miR-199a-3p), possibly indicating that the two represented different developmental stages (Bantounas et al., 2021). This example demonstrates the main advantage of this method, which is the acquisition of spatial information regarding the expression of the miRNAs of interest within a given tissue. This information would be lost if one were to rely exclusively on transcriptional detection methods [e.g., qPCR or Next Generation miRNA Sequencing (miRseq)]. The hybridization steps of the protocol presented here (see below) were adapted from Jørgensen et al. (2010) and were optimized for kidney/kidney organoids and the particular miRNAs we studied (Bantounas et al., 2021). However, the method can in principle be used with formalin-fixed, paraffin-embedded (FFPE) sections of any tissue and for any miRNA or other small RNA of similar size.
Materials and Reagents
1.5 ml Eppendorf tubes (Starlab, catalog number: S1615-5550)
Plastic pipette tips (preferably filtered) (Starlab, catalog numbers: S1122-1830 [1,000 μl]; S1120-8810 [200 μl]; S1123-1810 [20 μl]; S1121-3810 [10 μl]; or equivalent)
Vacuum filter units, pore size 0.2 μm (Thermo, catalog number: 568-0020)
6-well plates (Costar, catalog number: 3516, or equivalent)
3 ml plastic Pasteur pipettes (Starlab, catalog number: E1414-0311, or equivalent)
Tissue processing/embedding cassettes (Simport, catalog number: M490-4) (Figure 1, Left)
Stainless steel or plastic tissue processing capsules (Figure 1, Right)
Note: The capsules depicted in Figure 1 were the ones used by the authors but have now been discontinued. Therefore, alternative options are proposed here: Simport, catalog number: M470; or Fisher, catalog number: 15-182-219; or equivalent.
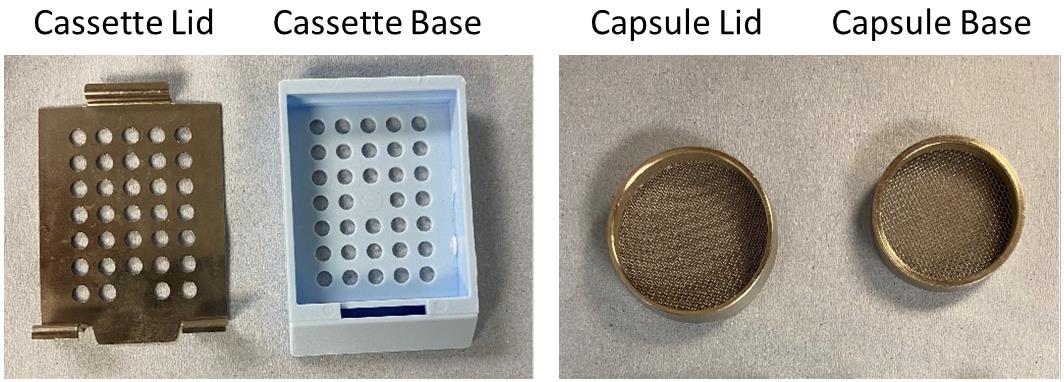
Figure 1. Tissue processing cassette and capsuleMetal molds for paraffin embedding of tissue (Leica, catalog number: 3803081E)
RNaseZap® wipes for disinfection of working surfaces (Invitrogen/Ambion, catalog number: AM9788)
Superfrost® Plus slides (ThermoFisher Scientific, catalog number: J1800AMNZ)
Human embryonic kidney tissue was provided by the MRC and Wellcome Trust Human Developmental Biology Resource (http://www.hdbr.org/)
hPSC-derived kidney organoids can be produced according to published protocols (Takasato et al., 2015; Bantounas et al., 2018 and 2021). In our study (Bantounas et al., 2021), we differentiated the MAN13 human embryonic stem cell (hESC) line (Ye et al., 2017), but any hESC or induced pluripotent stem cell (iPSC) line can be used in principle
Paraffin wax for embedding tissue, melting point 61°C (Pfm Medical, catalog number: 9000)
Nuclease-free water (not DEPC-treated) (ThermoFisher Scientific, catalog number: AM9930)
Phosphate buffered saline (PBS), without Ca2+ and Mg2+ (Sigma, catalog number: D8537-500 ml)
5’-Digoxigenin-labelled miRCURY® LNA® miRNA detection probes (1 nmol) (see Note 1) (QIAGEN, catalog number: 339111; see below for individual probe codes). In this example, we used probes targeting:
miR-199a-3p (QIAGEN, catalog number: YD00615410)
miR-214-3p (QIAGEN, catalog number: YD00611471)
scrambled probe (negative control; QIAGEN, catalog number: YD00699004)
U6 snRNA (positive/optimization control; QIAGEN, catalog number: YD00699002)
miRCURY® LNA® miRNA ISH Buffer Set (FFPE) (QIAGEN, catalog number: 339450), which includes:
2× Formamide-free miRNA ISH buffer
Proteinase K solution
UltraPureTM 20× SSC Buffer (Invitrogen, catalog number: 15557-044)
Tween-20 (Sigma, catalog number: P1379-250ML)
Sheep anti-DIG-AP (alkaline phosphatase linked) antibody (Roche/Sigma-Aldrich, catalog number: 11093274910)
Sheep serum (Sigma-Aldrich, catalog number: S3772-10ML)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A1470-100G)
Nitro blue tetrazolium/5-bromo-4-chloro-indolyl-phosphate (NBT/BCIP) ready-to-use tablets (Roche, catalog number: 1 1 697 471 001)
Levamisol (Fluka, catalog number: 31742), diluted in water to make a 100 mM stock solution
Nuclear Fast Red solution (Merck, catalog number: N3020)
Industrial methylated spirit (IMS) (various suppliers)
Xylene (various suppliers)
100% Ethanol (various suppliers)
KCl (various suppliers)
KP Marker Plus (Histolab, catalog number: 98307-R) or similar hydrophobic marking pen (various suppliers)
4% Paraformaldehyde (PFA) Solution (see Recipes)
1 M Tris Buffer (see Recipes)
0.5 M EDTA Solution (see Recipes)
5 M NaCl Solution (see Recipes)
1× Proteinase K Buffer (see Recipes)
SSC Buffer dilutions (see Recipes)
PBS-T 0.1% (see Recipes)
Blocking solution (see Recipes)
Antibody dilution buffer (see Recipes)
KTBT Buffer (see Recipes)
Equipment
Tissue Processor (Leica, model: ASP300S; or equivalent)
Heated Paraffin Embedding Station (Leica HistoCore Arcadia H or equivalent)
Cold Plate (Leica, HistoCore Arcadia C or equivalent)
Microtome (Leica, model: RM225; or equivalent)
Hybridization oven or other variable temperature incubator (e.g., Hybaid, Midi Dual 14 or equivalent)
Microscope slide rack (Simport, catalog number: M905-12DGY)
Jars for immersing the slide rack into solvents/buffers (Simport, catalog number: M900-12G)
Heating block (up to 90°C) (Eppendorf ThermoMixer F1.5 or equivalent)
Microcentrifuge (Labnet, Prism or equivalent)
Gilson® or equivalent pipettes (various suppliers)
Staining tray for holding the slides during hybridization and staining (see Figure 2).
Note: The tray depicted in Figure 2 was the one used by the authors but has now been discontinued. An alternative is proposed here: Fisher, catalog number: 22-045-035.
Waterbath (Grant SUB6, catalog number: P266; or equivalent)
Autoclave (various suppliers)
Optionally, for imaging: 3D-Histech Panoramic-250 microscope slide-scanner, with a 40×/0.95 Plan Apochromat objective (Zeiss)
Software
Optional: CaseViewer (3DHISTECH Ltd.; www.3dhistech.com), to capture images following scanning with a 3D-Histech slide-scanner (see point 14, in Equipment)
Fiji/ImageJ (http://imagej.net/Fiji/Downloads) (for image processing following capture)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Lopes, F. M., Kimber, S. J. and Bantounas, I. (2021). In situ Hybridization of miRNAs in Human Embryonic Kidney and Human Pluripotent Stem Cell-derived Kidney Organoids. Bio-protocol 11(17): e4150. DOI: 10.21769/BioProtoc.4150.
分类
干细胞 > 多能干细胞 > 再生医学
发育生物学 > 细胞生长和命运决定
分子生物学 > RNA > RNA 标记
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link