- EN - English
- CN - 中文
Cytoduction and Plasmiduction in Yeast
酵母细胞诱导中的细胞诱导和质粒诱导
发布: 2021年09月05日第11卷第17期 DOI: 10.21769/BioProtoc.4146 浏览次数: 3675
评审: Emilia KrypotouLucy XieIndranil MalikAnonymous reviewer(s)
Abstract
Cytoduction, and a related technique referred to as plasmiduction, have facilitated substantial advancements in the field of yeast prion biology by providing a streamlined method of transferring prions from one yeast strain to another. Prions are cytoplasmic elements consisting of aggregated misfolded proteins, and as such, they exhibit non-Mendelian patterns of inheritance. While prion transfer through mating and sporulation, or through protein transformation, is possible, these approaches yield non-isogenic strains or are technically complex, respectively. Cytoduction is a mating-based technique that takes advantage of a kar1 mutation with impaired nuclear fusion (karyogamy). It is a straightforward method for introducing a prion to any yeast strain (referred to as the recipient) by mating it with a donor strain containing the prion of interest. The only absolute requirement is that one of these two strains (donor or recipient) must carry the kar1-1 mutation to limit nuclear fusion. The resulting cytoductant contains the original nucleus of the recipient strain, but a cytoplasm reflecting a mix of all elements from the donor and the recipient. Modifications to the basic cytoduction strategy provide several options for successful cytoduction, including when working with slow growing or respiratory deficient strains. A significant advantage of the plasmiduction protocol presented is the ability to transfer a plasmid encoding a fluorescently tagged version of the prion protein, which allows for the direct verification of the prion state through visual protein aggregates.
Graphic abstract:
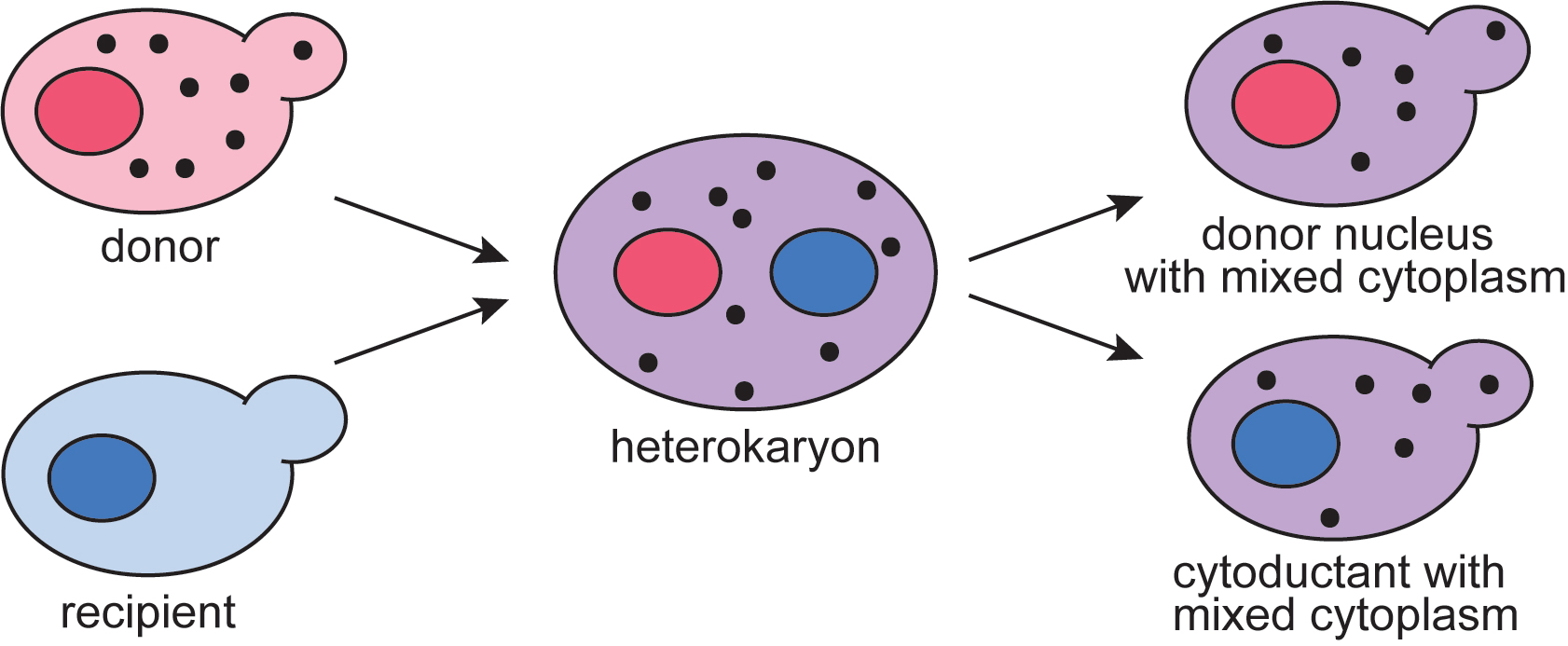
Transfer of Yeast Cytoplasmic Elements such as Prions using Cytoduction
Background
Cytoduction is a technique developed in yeast during the 1970s to study cytoplasmic inheritance (Conde and Fink, 1976; Zakharov and Yarovoy, 1977). Cytoduction takes advantage of a karyogamy mutant, kar1-1, in which nuclear fusion during mating is limited (Conde and Fink, 1976; Dutcher, 1981; Livingston, 1977). In a mating between KAR1 and kar1-1 cells, the cytoplasm is mixed but two distinct nuclei are maintained (Figure 1). The resulting daughter cells from this heterokaryon contain a mix of cytoplasm from both parents, yet only one of the original parental nuclei. This technique has been used as a tool to study and track other cytoplasmic elements such as mitochondria (Lancashire and Mattoon, 1979), killer toxins (Seki et al., 1985), and oligomycin resistance (Matsuoka et al., 1982). Cytoduction can also be used to replenish rho0, or mitochondrial DNA deficient, strains with fresh mitochondria, thus restoring respiration deficiencies associated with the loss of mitochondrial DNA (Merz and Westermann, 2009; Stenger et al., 2020). Because many terms associated with cytoduction may not be universally familiar, definitions are available for reference in Table 1.
Cytoduction has also been extremely helpful in the study of yeast prions. Yeast prions exhibit non-mendelian inheritance (Cox, 1965; Young and Cox, 1972), in which traditional mating and sporulation show a 4:0 pattern of inheritance. Cytoduction was used to show that several prions, including [PSI+], can be inherited through cytoplasmic transfer (Cox et al., 1980; Derkatch et al., 2001; Brachmann et al., 2005). Since these early studies, cytoduction and several other variant protocols have not only provided solid evidence that prions are cytoplasmic elements, but have facilitated important insight into the genetic mechanisms underlying prion formation (Masison et al., 1997; Manogaran et al., 2011) and how prions are maintained in populations (Manogaran et al., 2010; Bateman and Wickner, 2013; Keefer and True, 2016; Dorweiler et al., 2020).
Despite impaired nuclear fusion in kar1-1/KAR1 heterokaryons, rare exchange of genetic information between nuclei occurs at a frequency that is inversely proportional to the size of the chromosome (Dutcher, 1981). To circumvent this downside of unintentional chromosomal transfer to cytoductants, the Rothstein lab has developed a universal donor strain in which each chromosome contains a galactose-inducible conditional centromere that can be destabilized after mating (Manogaran et al., 2010; Reid et al., 2011). Even if a rogue chromosome is transferred into the recipient strain, growth on galactose containing media results in chromosome destabilization and loss.
Plasmiduction is a closely related process that uses the principles of cytoduction and the rare exchange of nuclear information to transfer plasmids between strains. Originally used to transfer plasmids into strains that were difficult to transform, plasmiduction can be used to transfer centromeric, 2 μ, and yeast artificial chromosomes into recipient strains (Brown et al., 1981; Natsoulis et al., 1994; Spencer et al., 1994). While the ability to transfer extrachromosomal DNA between strains is a valuable tool of its own right, when transferring prions between strains, plasmiduction offers the ability to include additional plasmid-associated selectable markers to help identify cytoductants. Additionally, plasmiduction can circumvent the need for subsequent plasmid transfer into cytoductants as part of the prion verification process.
Here, we present three different cytoduction strategies. We begin by presenting the classic protocol that relies upon using a cycloheximide resistant, rho0 recipient strain, and provides the most direct selection of cytoductants. Despite this significant advantage, this classic protocol is not always the most appropriate for growth sensitive or respiratory deficient strains. We also present two additional cytoduction strategies for generating cytoductants, including plasmiduction.
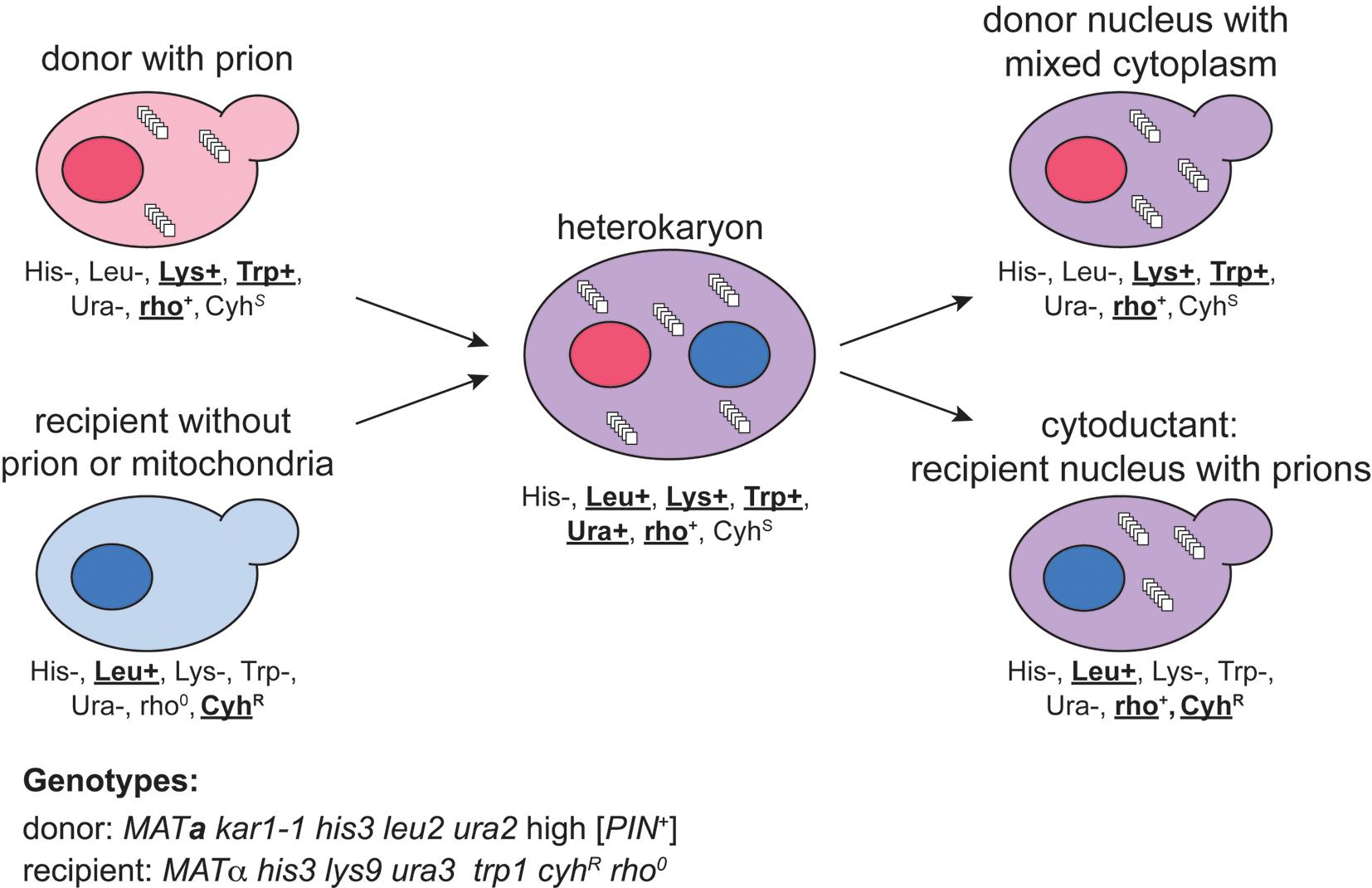
Figure 1. Schematic example of cytoduction. A donor yeast strain carrying the prion of interest is mated with a recipient strain that lacks the prion. Successful mating only happens when the two strains are of opposite mating type (MATa vs. MATα), and fusion between the two nuclei is limited only if one of the original strains carries the karyogamy deficient kar1-1 mutation. Using the specific example genotypes provided for the donor and recipient, phenotypic labels are assigned to all cells within the schematic, with wild type or cycloheximide resistance phenotypes in bold. This practice reveals those phenotypes that can be used to select the desired cytoductants from all other cells in the process. Note that donor (ura2) and recipient (ura3) contain mutations in different genes within the uracil biosynthesis pathway. The heterokaryon is heterozygous for each of these genes, and therefore Ura+ confers growth on SD-Ura media.
Table 1. Terminology associated with prion cytoduction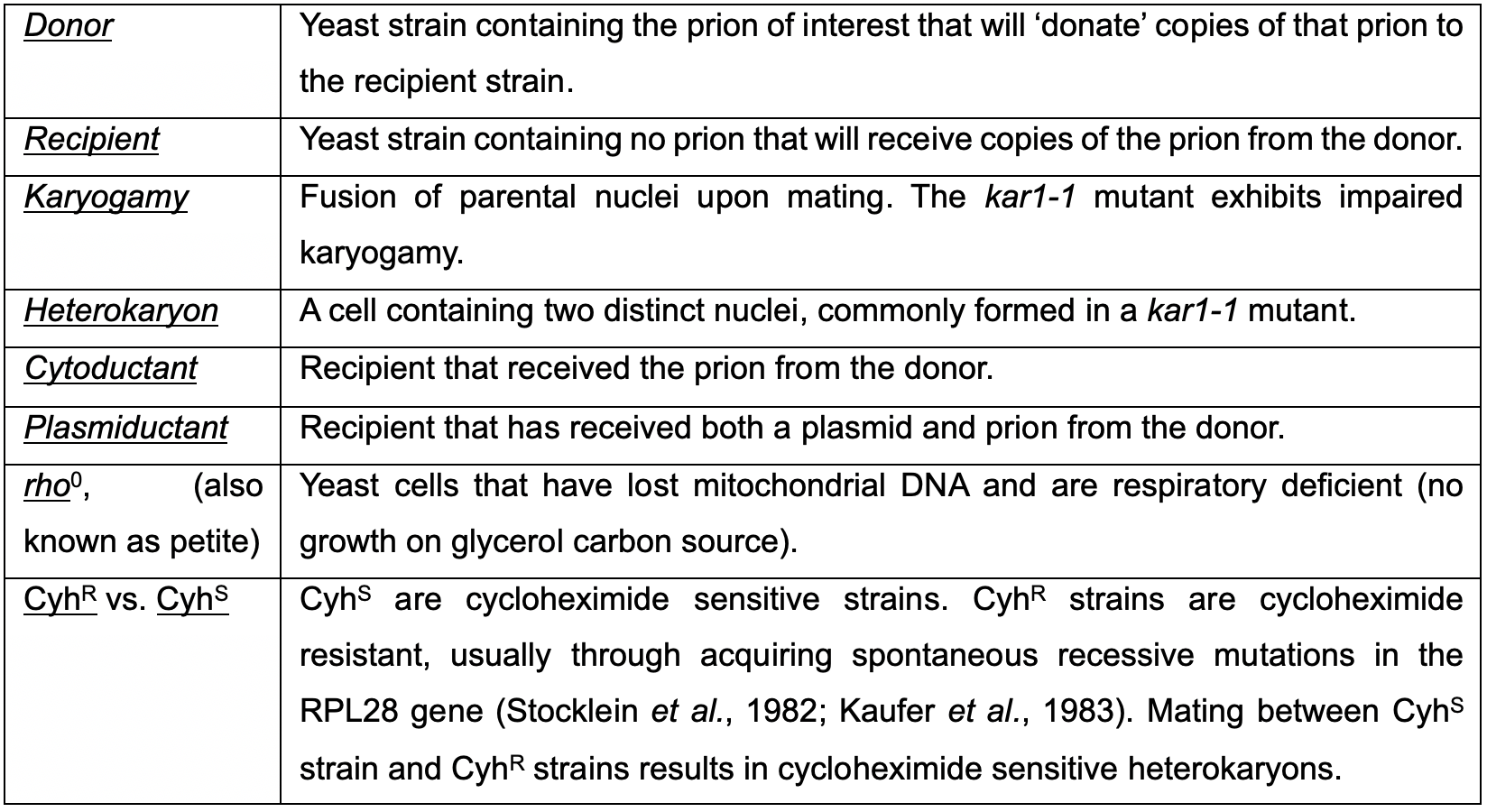
Materials and Reagents
Petri Dishes with Clear Lid (FisherbrandTM, catalog number: FB0875712)
Toothpicks (flat, sterilized)
Slides and coverslips (e.g., Fisherbrand, catalog numbers: 12-550-A3 and 12-542A)
Yeast strains
Donor and recipient strains of complementary mating type; MATa and MATα.
One of these, ideally the donor, must contain the kar1-1 mutant allele.
Unique auxotrophic or antibiotic markers that differentiate donor and recipient strains to significantly streamline isolation and confirmation of cytoductants.
To use the plasmiduction protocol described in Section D below, the donor and recipient strains should share at least one auxotrophic marker that will be complemented by the plasmid. Alternatively, a plasmid conferring antibiotic resistance (such as G418 resistance) could be used.
One additional yeast strain (referred to generically throughout this protocol as your “tester strain,” for instance BY4741, BY4742, or another favorite strain; it must meet the following conditions).
Must be of opposite mating type relative to your recipient strain.
Should be wild type for the KAR1 gene.
Should include unique auxotrophic markers relative to your recipient strain to facilitate diploid selection.
Use of this strain to screen cytoductants for fluorescent aggregates (as described in Step B4a) demands two additional requirements. First, the strain must share at least one auxotrophic marker relative to your recipient strain that facilitates maintenance of a plasmid necessary to confirm prion status of your cytoductants (as outlined below in Step B4a). For example, if using a URA3 plasmid, both your recipient and tester strain must be ura3. Second, the strain should be devoid of all prions, but minimally must be devoid of the prion being cytoduced. If prion status of this tester strain is uncertain, cure the strain using guanidine hydrochloride (as described below in Step B4b). Regardless of whether you suspect that the tester strain is free of prions or have cured the strain yourself, it is prudent to verify that the strain contains no prions by visualizing cells via fluorescent microscopy as described (Step B4a, focusing on parts i., and v.-vii. only, as mating will not be required). If properly cured, these cells will have diffuse fluorescence under the conditions described in Step B4a.
Note: This protocol focuses predominately on Hsp104-dependent prions and thus describes the guanidine hydrochloride method for curing these prions. If you are working with prions that require an alternative curing method, such as [GAR+] or [SMAUG+], then curing of your tester strain should be completed using your prion specific curing method rather than that described in Step B4b.
Plasmids for prion identification or plasmiduction
To detect [PSI+] using fluorescence microscopy, plasmid with fluorescently tagged Sup35 protein (e.g., pCUP1-Sup35-GFP, with URA3, Addgene.org #1087)
To detect [PIN+] using fluorescence microscopy, plasmid with fluorescently tagged Rnq1 protein (e.g., pCUP1-Rnq1-YFP, with URA3, Addgene.org #15596)
Plasmids that provide optional antibiotic or auxotrophic selection (for the plasmiduction protocol).
Media/Agar plates
Dehydrated Agar for solid media (BactoTM, catalog number: 214030; 2 kg, or smaller quantity)
YPD (ingredients, recipes in separate section below)
Yeast Extract, low-dusting (LD) (GibcoTM, DifcoTM, catalog number: 210933)
Peptone (GibcoTM, BactoTM, catalog number: 211677)
Dextrose Glucose, Anhydrous, 2 kg (BDTM DifcoTM, catalog number: 215510 or smaller quantity)
Components for Synthetic Dropout plates
Yeast Nitrogen Base without Amino Acids and Ammonium Sulfate (DifcoTM, catalog number: 233520)
Ammonium sulfate, 99.5%, for analysis (ACROS OrganicsTM, catalog number: 205870010)
Nutrients for dropout powders (for an exhaustive list, including relative proportions, see Sherman, 2002)
1)Amino acids: L-arginine-HCl, L-histidine, L-leucine, L-lysine mono-HCl, L-methionine, L-phenylalanine, L-tryptophan, L-tyrosine, L-valine (Sigma-Aldrich)2)Adenine hemisulfate salt (Sigma-Aldrich, catalog number: A-9126)3)Uracil (Sigma-Aldrich, catalog number: U-0750)4)Some yeast strains may have additional unique auxotrophic markers and require corresponding nutrients (Sherman, 2002).
Cycloheximide solution (for detailed protocol B only, see Recipes)
Cycloheximide (Sigma, catalog number: C7698)
DMSO (Sigma, catalog number: D8418)
Ethidium bromide (if recipient strain needs to be rendered rho0 for appropriate cytoduction strategy (Sigma, catalog number: E8751)
Glycerol (Sigma, catalog number: G-5516)
Possibly one or more yeast antibiotics dependent upon cytoductant selection strategy. Two common yeast antibiotics are geneticin (Sigma, catalog number: A-1720; Winzeler et al., 1999) and nourseothricin (GoldBio, catalog number: N-500-1; Haarer et al., 2007).
Glass beads for spreading yeast cells (Zymo Research, catalog number: S1001)
Reagents for yeast transformation (Gietz, 2014)
ssDNA (Sigma, catalog number: D7656)
PEG 3350 (Sigma, catalog number: 88276)
Tris (C4H11NO3, GoldBio, catalog number: T-400-1)
EDTA (EMD, catalog number: EX0539-1)
Lithium Acetate (Sigma, catalog number: L4158)
Guanidine Hydrochloride (Sigma, catalog number: G4505)
Copper Sulfate (Sigma, catalog number: C1297)
Media (see Recipes)
10 mg/ml Cycloheximide stock solution (see Recipes)
YPD (or YPGlycerol) plates with 10 μg/ml Cycloheximide (see Recipes)
2.5 mg/ml Ethidium Bromide (ETBr; see Recipes)
YPD plates with 25 μg/ml EtBr (see Recipes)
5 M Guanidine Hydrochloride (GuHCl; see Recipes)
YPD plates with 5 mM GuHCl (see Recipes)
SGlycerol-selective plates (see Recipes)
Equipment
Fluorescent microscope (to monitor the presence of prion via fluorescent tag)
Replica plater/velveteen squares (Fisher Scientific, catalog number: 09-718-1)
Autoclave
Incubator (30°C)
Erlenmeyer Flasks (1 L for media preparation)
(All equipment is available from any standard scientific supply distributor; specific brands/models are not required)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Dorweiler, J. E. and Manogaran, A. L. (2021). Cytoduction and Plasmiduction in Yeast. Bio-protocol 11(17): e4146. DOI: 10.21769/BioProtoc.4146.
分类
细胞生物学 > 细胞工程
生物科学 > 生物技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link