- EN - English
- CN - 中文
Agrobacterium-mediated Transformation of Japonica Rice Using Mature Embryos and Regenerated Transgenic Plants
农杆菌介导粳稻成熟胚和再生转基因植株的转化
发布: 2021年09月20日第11卷第18期 DOI: 10.21769/BioProtoc.4143 浏览次数: 6179
评审: Bedabrata SahaAnonymous reviewer(s)
Abstract
Identification of novel genes and their functions in rice is a critical step to improve economic traits. Agrobacterium tumefaciens-mediated transformation is a proven method in many laboratories and widely adopted for genetic engineering in rice. However, the efficiency of gene transfer by Agrobacterium in rice is low, particularly among japonica and indica varieties. In this protocol, we elucidate a rapid and highly efficient protocol to transform and regenerate transgenic rice plants through important key features of Agrobacterium transformation and standard regeneration media, especially enhancing culture conditions, timing, and growth hormones. With this protocol, transformed plantlets from the embryogenetic callus of the japonica cultivar ‘Taichung 65’ may be obtained within 90 days. This protocol may be used with other japonica rice varieties.
Keywords: Electroporation (电穿孔)Background
Genetic transformation and expression of recombinant proteins in plant cells are major factors for plant genetic engineering. This is also a powerful tool for discovering novel genes and exploring genetically controlled traits. Agrobacterium tumefaciens-mediated transformation has been known for its unique ability to transfer a DNA segment from a specialized plasmid into a host plant cell (Gelvin, 2010). This feature allows efficient insertion of stable, unrearranged, single-copy DNA into plant genomes, which may lead to more stable expression than multiple gene copies or scrambled inserts (Iglesias et al., 1997). Since the initial reports in the early 1980s using Agrobacterium to generate transgenic plants, efforts were made to improve the tool in plant transformation. In most instances, major improvements involved alterations in plant tissue culture transformation and regeneration conditions rather than manipulation of host or bacterial genes. The first effective method for Agrobacterium-mediated transformation of japonica rice is now a common protocol in many laboratories (Chan et al., 1993; Hiei et al., 1994; Park et al., 1996). Many functional analyses, for example, PCR, GUS assay, and southern blot analysis, have confirmed the integration of foreign genes into transgenic rice plants obtained by Agrobacterium-mediated transformation (Dai et al., 2001); moreover, Mendelian inheritance of the transgenes was also reported (Pawlowski and Somers, 1996; Hiei et al., 1997).
The transfer of T-DNA and its integration into the rice genome is influenced by numerous factors, such as genotype, explants, Agrobacterium strains, plasmid vectors, addition of vir-gene inducing synthetic phenolic compounds, selection marker, and various conditions of tissue culture and regeneration. In tissue culture systems, selection of actively growing regenerable calli is a principal factor for efficient plant transformation. Likewise, optimization of culture conditions for co-cultivation of rice calli with Agrobacterium (Ozawa and Takaiwa, 2010) and suppression of Agrobacterium overgrowth may be applied to improve the transformation efficiency. Scientists have attempted to improve the plasmid constructions and rice transformation by Agrobacterium-mediated transformation to investigate the function of seed storage protein gene factors as well as cloning genomic candidate regions of more than 10 kb, which may have low restriction enzymes sites required for cloning. In particular, previous limitations of efficient promoter expression were overcome (Gupta et al., 2001; Furtado et al., 2008), especially for endosperm-specific expression (Zhou et al., 2013).
Several transformation methods have been established by many laboratories using mature japonica and indica seeds. Taichung 65 is a japonica rice variety, selected from a cross between the two Japanese varieties, Shinriki and Kameji, in 1923 (Iso, 1957). Taichung 65 is considered an important cultivar for studying rice genetics and breeding. Several point mutations were induced to the genetic stock of Taichung 65 using the methylnitrosourea (MNU) mutagen and have been maintained at the institute of plant genetic resources of Kyushu university. Taichung 65 has been extensively used as a model cultivar for rice biology, breeding research, and genomic studies initiated in many laboratories. However, transformation efficiency is still low in most indica and many japonica varieties due to the difficult regeneration; consequently, the procedure to obtain transgenic plants takes an average of 5 months (Nishimura et al., 2006). Thus, additional improvements in the transformation potentiality are still possible.
Here, we describe a protocol for Agrobacterium-mediated transformation in rice using mature embryos. This protocol is based on the method described by Toki (Toki, 1997), with several modifications; in particular, meropenem is used for plant regeneration (Ogawa and Mii, 2007). Meropenem exhibits the highest antibacterial activity and also achieves a high shoot formation rate in rice and tomato (Ogawa and Mii, 2007). Using this protocol, we succeeded to produce hundreds of independent transgenic lines and transformed several novel genes over the past decade, including Endosperm Storage Protein (ESP1), encoding a eukaryotic chain release factor1 (eRF1) (Elakhdar et al., 2019); Esp2, encoding the protein disulfate isomerase 1-1 (PDI 1-1) (Satoh-Cruz et al., 2010); Glup3, encoding a vacuole processing enzyme (VPE) (Kumamaru et al., 2010); Glup4, encoding the small GTPase Rab5a (Fukuda et al., 2011); and Glup6, encoding the guanine nucleotide exchange factor (GEF) (Fukuda et al., 2013).
In summary, this protocol is a step-by-step approach to obtain stably transformed plants from mature rice embryos by optimizing several stages of Agrobacterium transformation and standard regeneration medium components, including culture conditions, timing, and growth hormones. In this method, we use a 13.8 kb construct carrying the hygromycin phosphotransferase gene (hpt), a firefly luciferase reporter gene (LUC), spectinomycin (Sp), an attR1 site, the chloramphenicol resistance gene (CmR), the ccdB gene, the attR2 site, and the ZmUbi1 promoter (Figure 1 and Figure 2).

Figure 1. Flow chart for Agrobacterium-mediated transformation and regeneration of rice calli using mature embryogenic seed (cv; Taichung 65). (a) Seed embryos. (b) Calli pieces with granular structure, yellowish-white. (c) Growth of transformed Agrobacterium harboring the pSMAH638OX/Ubilp binary vector. (d) Co-cultivation of infected calli. (e) De-colonization of Agrobacterium tumefaciens. (f) Shoot regeneration. (g) Rooting in regenerated shoots. (h) Hardening of rooted plants. (i) Transplanted hardened plants.
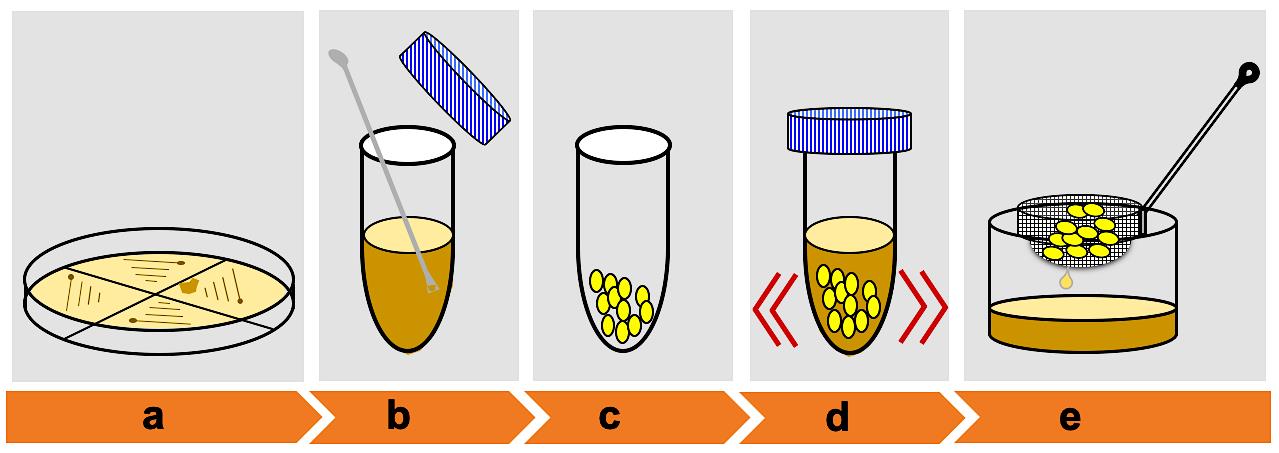
Figure 2. Schematic showing embryo calli infection by Agrobacterium. (a) Growth of Agrobacterium-mediated harboring pSMAH638OX/Ubilp vector on LB medium. (b) Mix a small portion of the positive colonies. (c) Collect healthy calli for infection. (d) Calli infection. (e) Remove excess Agrobacterium suspension.
Materials and Reagents
Plasticware and Glass
Petri dish, 90 × 20 mm
Petri dish, 90 × 15 mm
24-well multi-well plates
Sterile plastic falcon tubes, 50 ml
Filter paper, 70 mm
Parafilm tape (Parafilm Pechiney PM996; 125' L X 4"; www.parafilm.com)
Sterile syringe filters 0.22 μm
Sterile syringe, 30 ml
Stainless-steel sieve
Microspatula
Plant culture pots for rooting (6.5 × 6.5 cm)
Plant soil pots (23 × 23 × 19 cm)
Biological materials
Mature dry seeds of japonica cv. Taichung 65
The target cultivar of the current transformation method.
Binary vector containing gene of interest
In the following protocol, the binary vector pSMAH638OX/Ubilp (13.8 Kb) construct was derived by the Gateway recombination cloning technology (InvitrogenTM, Thermo Fisher Scientific, Inc.), harboring firefly luciferase (LUC) reporter gene, the hygromycin phosphotransferase (hpt) selectable gene marker, spectinomycin (Sp), an attR1 site, the chloramphenicol resistance gene (CmR), the ccdB gene, and the attR2 site. We used the ZmUbi1 promoter for LUC expression (Hakata et al., 2010; Elakhdar et al., 2019)
Agrobacterium tumefaciens strains EHA101 or EH105 (Hood et al., 1993).
Reagents and chemicals
Ethanol CH3CH2OH (Sigma-Aldrich, catalog number: N0640), 70% (v/v) in distilled water, store at room temperature (RT)
Sodium hypochlorite 10% NaClO (Sigma-Aldrich, catalog number: 105614), store at RT
Sodium chloride, NaCl (Sigma-Aldrich, catalog number: 7647-14-5), store at RT
Sodium hydroxide, NaOH (Sigma-Aldrich, catalog number: 1310-73-2), store at RT
Tween® 20 EMD, (CALBIOCHEM, catalog number: 655205), store at RT
Sucrose C12H22O11 (Wako, catalog number: 196-00015), store at RT
D-Glucose C6H12O6 (Sigma-Aldrich, catalog number: 07-0680-5), store at RT
D-Sorbitol C16H14O6 (Sigma-Aldrich, catalog number: 28-4770-5), store at RT
Casamino acid (Sigma-Alorich, catalog number: 65072-00-6), store at RT in dry condition
L-Proline (PEPTIOE, catalog number: 2718), store at RT
CHU (N6) Basal salt mixture (Sigma-Aldrich, catalog number: C1416-10L), store at 2-8°C
Murashige & Skoog Basal Medium (Sigma-Aldrich, catalog number: M552450L), store at 2-8°C
CHU N6-Vitamin solution X1000 (Phyto, catalog number: PHT:C149), store at 2-6°C
LB Broth, Miller (Luria-Bertani; BD Difco, catalog number: 244620), store at RT
Bacto Peptone (BD Difco, catalog number: 211677), store at RT
Bacteriological agar (Bacto Agar; BD Difco, catalog number: 214010), store at RT
2,4-Dichlorophenoxyacetic acid (Wako, catalog number: 040-18532), store at RT
1-Naphthaleneacetic acid NAA C10H7CH2COOH (Wako, catalog number: 86-87-3), store at RT
Kinetin C10H9N5O (Wako, catalog number: 110-00331), store at 2-10°C
Gelrite (0.4%) (Wako, catalog number: 067-04035), store at RT
Carbenicillin sodium salt (Sigma-Aldrich, catalog number: C1389-5G), store at 2-8°C
Hygromycin B (Wako, catalog number: 085-06153), store at -20°C
Acetosyringone (Sigma-Aldrich, catalog number: D134406-5G), store at -20°C
Meropenem (Dainippon Sumitomo Pharma, Osaka, Japan), store at -20°C
Dimethyl sulfoxide DMSO (CH3)2SO (Wako, catalog number: 046-21981), store at RT
KOD-FX (TOYOBO, catalog number: KFX-101), store at -20°C
Sterilizing solution (see Recipes)
Solution A
Solution B
Antibiotics (see Recipes)
Carbenicillin (500 mg/ml)
Hygromycin (50 mg/ml)
Kanamycin (50 mg/ml)
Acetosyringone (100 mg/ml)
Acetosyringone (10 mg/ml)
Meropenem (12.5 mg/ml)
2,4-D (0.2 mg/ml)
NAA (0.2 mg/ml)
Kinetin (1 mg/ml)
Cultivation medium (see Recipes; Table 1)
LB sold medium
LB liquid medium
YEP medium
Notes:
All liquids and equipment containing Agrobacterium must be appropriately sterilized.
Essential elements that may influence the effectiveness of transformation are reagents and chemicals. Therefore, we strongly suggest following the methodology, particularly when starting a new protocol. The usage and storage conditions of chemicals are different; therefore, please make sure to follow the recommendations of each company, with particular attention to hazard information on the substances.
Equipment
-86°C Ultra-Low temperature freezer (SANYO, catalog number: MDF-U538)
-30°C Ultra-Low temperature freezer (SANYO, catalog number: MDF-792AT)
MicroPluserTM (Bio-Rad, Hercules)
Spectrophotometer (Thermo ScientificTM, model: NanoDrop 2000)
Incubator/ shaker 28°C for Agrobacterium
Plant growth chamber (SANYO)
Shaker for seed sterilization
Laminar flow hood (SANYO)
Autoclave
pH meter
Spinbar® magnetic stir bar (Sigma-Aldrich, catalog number: Z127116)
Polycarbonate vacuum desiccator (SANSYO, catalog number: SPD-WVGT240)
Thermal cycler (Bio-Rad, model: Tetrad 2 Thermal Cycler (4 × 96-well)
Locking system greenhouse
Rice seedling raising soil (JA Kumiai King Soil; Agr. Japan Co., Ltd.)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Elakhdar, A., Fukuda, M. and Kubo, T. (2021). Agrobacterium-mediated Transformation of Japonica Rice Using Mature Embryos and Regenerated Transgenic Plants. Bio-protocol 11(18): e4143. DOI: 10.21769/BioProtoc.4143.
分类
植物科学 > 植物转化 > 农杆菌介导的转化方法
环境生物学 > 植物
分子生物学 > DNA > DNA 克隆
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link