- EN - English
- CN - 中文
Using Atomic Force Microscopy to Study the Real Time Dynamics of DNA Unwinding by Mitochondrial Twinkle Helicase
使用原子力显微镜研究线粒体闪烁解旋酶对DNA解旋的实时动力学
发布: 2021年09月05日第11卷第17期 DOI: 10.21769/BioProtoc.4139 浏览次数: 3852
评审: Marc-Antoine SaniAntony ChettoorAbhilash Padavannil
Abstract
Understanding the structure and dynamics of DNA-protein interactions during DNA replication is crucial for elucidating the origins of disorders arising from its dysfunction. In this study, we employed Atomic Force Microscopy as a single-molecule imaging tool to examine the mitochondrial DNA helicase Twinkle and its interactions with DNA. We used imaging in air and time-lapse imaging in liquids to observe the DNA binding and unwinding activities of Twinkle hexamers at the single-molecule level. These procedures helped us visualize Twinkle loading onto and unloading from the DNA in the open-ring conformation. Using traditional methods, it has been shown that Twinkle is capable of unwinding dsDNA up to 20-55 bps. We found that the addition of mitochondrial single-stranded DNA binding protein (mtSSB) facilitates a 5-fold increase in the DNA unwinding rate for the Twinkle helicase. The protocols developed in this study provide new platforms to examine DNA replication and to explore the mechanism driving DNA deletion and human diseases.
Graphic abstract:
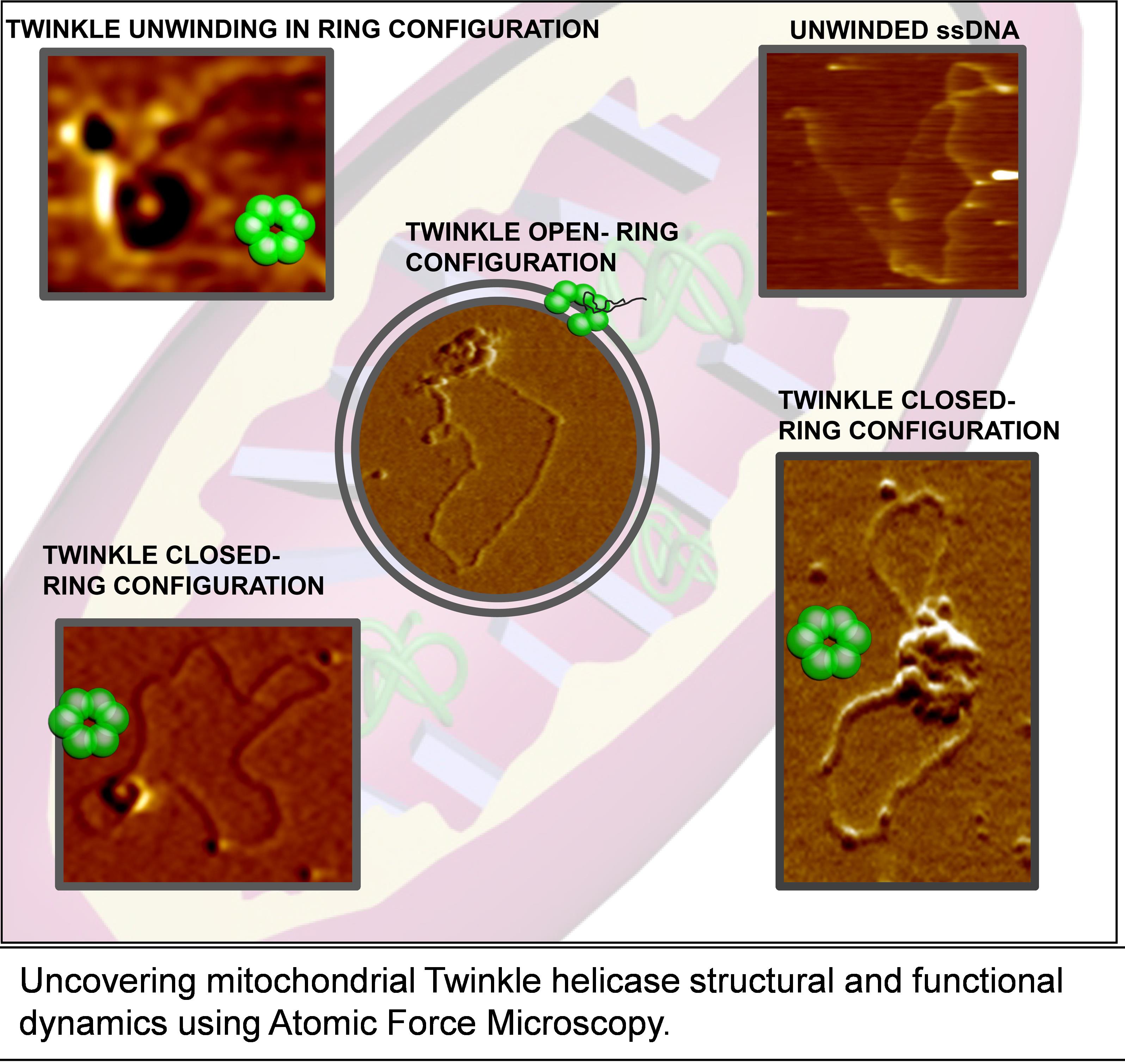
Mitochondrial Twinkle Helicase Dynamics
Background
Human mitochondria contain circular double-stranded DNA (16,569 bp) replicated by DNA polymerase Pol γ in concert with Twinkle helicase and mitochondrial single-stranded DNA binding protein (mtSSB) (Korhonen et al., 2004). Mutations in the genes encoding the core mitochondrial DNA (mtDNA) replication machinery have been associated with human diseases, including progressive external ophthalmoplegia, ataxia neuropathy syndromes, and other neurodegenerative diseases collectively known as “mitochondrial diseases” (Tyynismaa et al., 2005; Goffart et al., 2009; Ylikallio and Suomalainen, 2012; Bhatti et al., 2017; Suomalainen and Battersby 2018). The relationship between dysfunction in mtDNA replication and mitochondrial diseases makes it crucial to understand the molecular mechanism governing mtDNA replication. Although core mitochondrial proteins at the DNA replication fork have been identified (Bogenhagen et al., 2008), the structure and dynamics of mtDNA replication remain elusive. Most of the previous mitochondrial replication studies have been based on ensemble averaging methods using short DNA as template. The transient nature of the protein-DNA interactions presents significant challenges towards elucidating the underlying mechanisms using traditional methods. Using traditional methods, it has been shown that Twinkle is capable of unwinding dsDNA up to 20–55 bps both with and without mtSSB (Korhonen et al., 2004; Sen et al., 2012). To elucidate the mechanism of Twinkle helicase binding and unwinding DNA, we sought to uncover the intricate transient states, which may be obscured by ensemble averaging effects. To fill this knowledge gap, we developed protocols using Atomic Force Microscopy (AFM) imaging in air as well as in liquids that reveal a multi-faceted, dynamic structural description of DNA binding and unwinding by Twinkle-mtSSB at the single-molecule level. Studying unwinding reactions using AFM unveils unprecedented dynamic as well structural information of the unwinding reaction, helping us to provide a functional step model of Twinkle unwinding of the DNA. These protocols elucidated the real-time dynamics of Twinkle helicase loading onto DNA in an open ring configuration, switching to a closed ring state for DNA unwinding, and unloading from DNA as an open ring structure. These changes in the Twinkle helicase conformation dynamics are stochastic in nature and would have been lost in ensemble averaging traditional methods. Results from this study using this protocol showed that Twinkle helicase exists in various oligomeric states. Furthermore, time-lapse AFM imaging in liquids showed that Twinkle hexamers are formed through sequential recruitment of individual subunits, and these hexamers can exist and switch between both open and closed ring conformations. These imaging results support a model in which Twinkle helicase loads onto DNA in open-ring conformation and searches along DNA in a closed ring conformation. Also, the Twinkle helicase unwinds the DNA in open ring conformation and unloads from DNA upon reopening of the ring after performing DNA unwinding. Our AFM studies with a model circular dsDNA substrate containing 37-nt ssDNA gaps revealed that Twinkle is capable of unwinding thousands of base pairs when facilitated by mtSSB (~240 bp/min without mtSSB to ~1,265 bp/min with mtSSB) (Kaur et al., 2020). Here, we outline detailed protocols that can be adapted to study other DNA replication proteins in vitro using AFM imaging in air and time-lapse buffer imaging. While air imaging elucidates the spatial interaction, time-lapse buffer imaging provides dynamic information on protein-DNA interactions.
Materials and Reagents
100 mM ATP stock solution in deionized (DI) water buffered to pH 7.5 (aliquoted and stored at -80°C) (Sigma-Aldrich, catalog number: A7699)
Mica (SPI Chem Mica Grade V-1 50 × 25 mm × 0.15 mm, catalog number: 12001-26-2) (Figure 1)

Figure 1. SPI Chem Mica used for Atomic Force Microscopy (AFM) experimentsAminopropyl silatrane (APS) (Shlyakhtenko et al., 2013)
HEPES (Fisher Bioreagent, catalog number: 7365-45-9)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 7647-14-5)
Magnesium chloride (MgCl2) (Fisher Chemical, catalog number: 7791-18-6)
Glycerol (EMD Millipore, catalog number: 356350)
Nonidet P-40 (NP-40) (ThermoFisher Scientific, catalog number: 85124)
Nt.BstNBI (NEB, catalog number: R0607S)
Amicon Ultrafiltration (Millipore, catalog number: C82301, MW 100K)
ScaI restriction enzyme (NEB, catalog number: R3122S)
pscwo1 plasmid (Addgene, catalog number: 72300)
Helicase Imaging Buffer (see Recipes)
APS working solution (see Recipes)
Buffer 3.1: 1× Buffer components: NEB (see Recipes)
Equipment
NanoDrop 2000C (Thermo Scientific, catalog number: ND-2000C)
AFM tips for air imaging – Pointprobe PPP-FMR probes (Nanosensors, spring constant k = 2.8 N/m, Resonant frequency fr = 70 kHz).
AFM tips for Buffer imaging – BL-AC40TS (Biolever Mini) probe (Asylum Research, spring constant k = 0.09 N/m, Resonant Frequency fr = 110 kHz)
Centrifuge (to fit Eppendorf 1.5 ml tubes)
Cylinder of Nitrogen gas with a pressure regulator and nozzle.
Eppendorf tubes (1.5 ml) (Eppendorf, catalog number: 022431081)
Scotch Tape
MFP-3D – Bio AFM (Figures 2-4) (Asylum Research, Oxford Instruments)
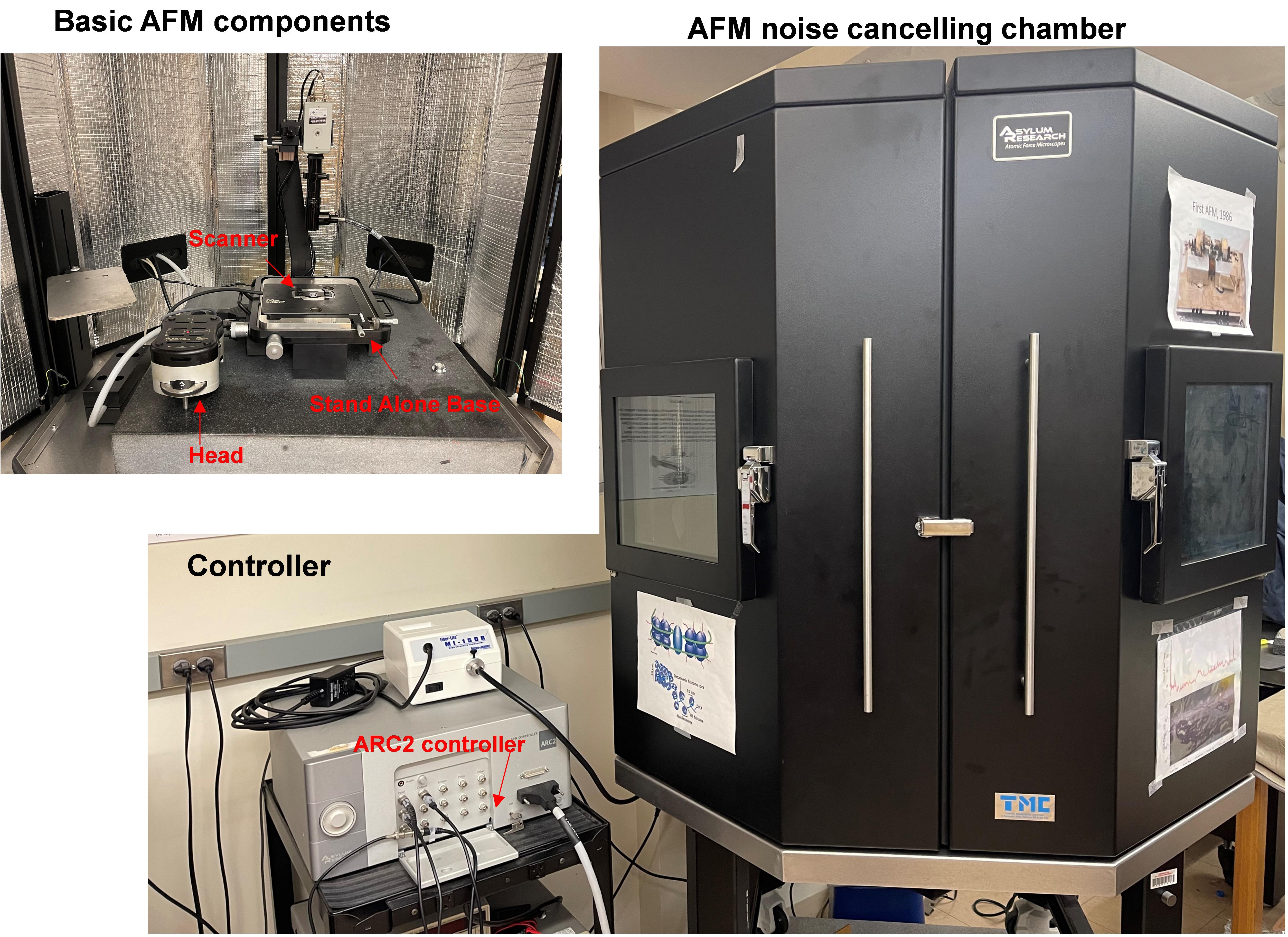
Figure 2. AFM instrument details showing basic AFM components inside the AFM noise canceling chamber with the AC2 controller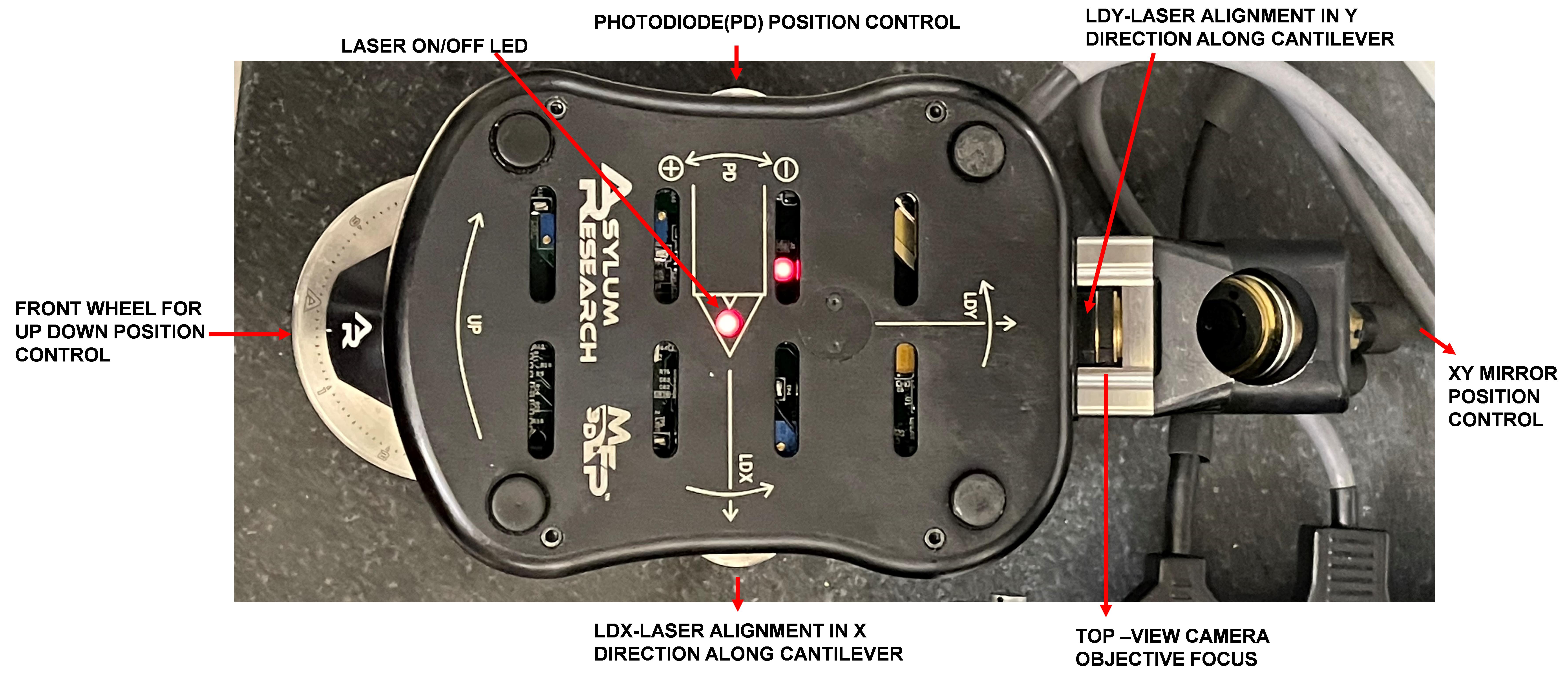
Figure 3. AFM head with labeled controls
Software
IGOR Pro 6 (https://www.wavemetrics.com/software/igor-pro-802, WaveMetrics, Portland, OR, USA)
MFP3D AFM imaging software or Gwydiion software (http://gwyddion.net/) (Nečas and Klapetek, 2012)
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kaur, P., Pan, H., Longley, M. J., Copeland, W. C. and Wang, H. (2021). Using Atomic Force Microscopy to Study the Real Time Dynamics of DNA Unwinding by Mitochondrial Twinkle Helicase. Bio-protocol 11(17): e4139. DOI: 10.21769/BioProtoc.4139.
- Kaur, P., Longley, M. J., Pan, H., Wang, W., Countryman, P., Wang, H. and Copeland, W. C. (2020). Single-molecule level structural dynamics of DNA unwinding by human mitochondrial Twinkle helicase.J Biol Chem 295(17): 5564-5576.
分类
生物物理学 > 显微技术 > 原子力显微镜
生物科学 > 生物技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link