- EN - English
- CN - 中文
Synchronized Real-time Measurement of Sec-mediated Protein Translocation
Sec介导的蛋白质易位的同步实时测量
发布: 2021年08月20日第11卷第16期 DOI: 10.21769/BioProtoc.4129 浏览次数: 3238
评审: Emilia KrypotouJose Antonio Reyes-DariasPetru-Iulian Trasnea
Abstract
The Sec translocon, consisting of a heterotrimeric transmembrane channel (SecYEG) and an associated ATPase (SecA), catalyzes the export of unfolded proteins from the cytosol in bacteria. Kinetically resolving protein translocation at high resolution yields mechanistic insight into the process. Translocation is typically followed by measuring the protection of proteins transported into lipid vesicles, which only allows visualization of translocation after it has already been completed and limits time resolution. Here, we describe the implementation of an assay for measuring translocation in real-time. By priming the reconstituted translocon with suitably engineered substrate proteins, the kinetics of the actual translocation process can be resolved at high resolution. To analyze translocation kinetics, we developed a detailed kinetic model of the process that includes on-pathway and off-pathway processes. Together, this experimental protocol and model permit detailed mechanistic analyses of Sec-dependent protein translocation.
Graphic abstract:
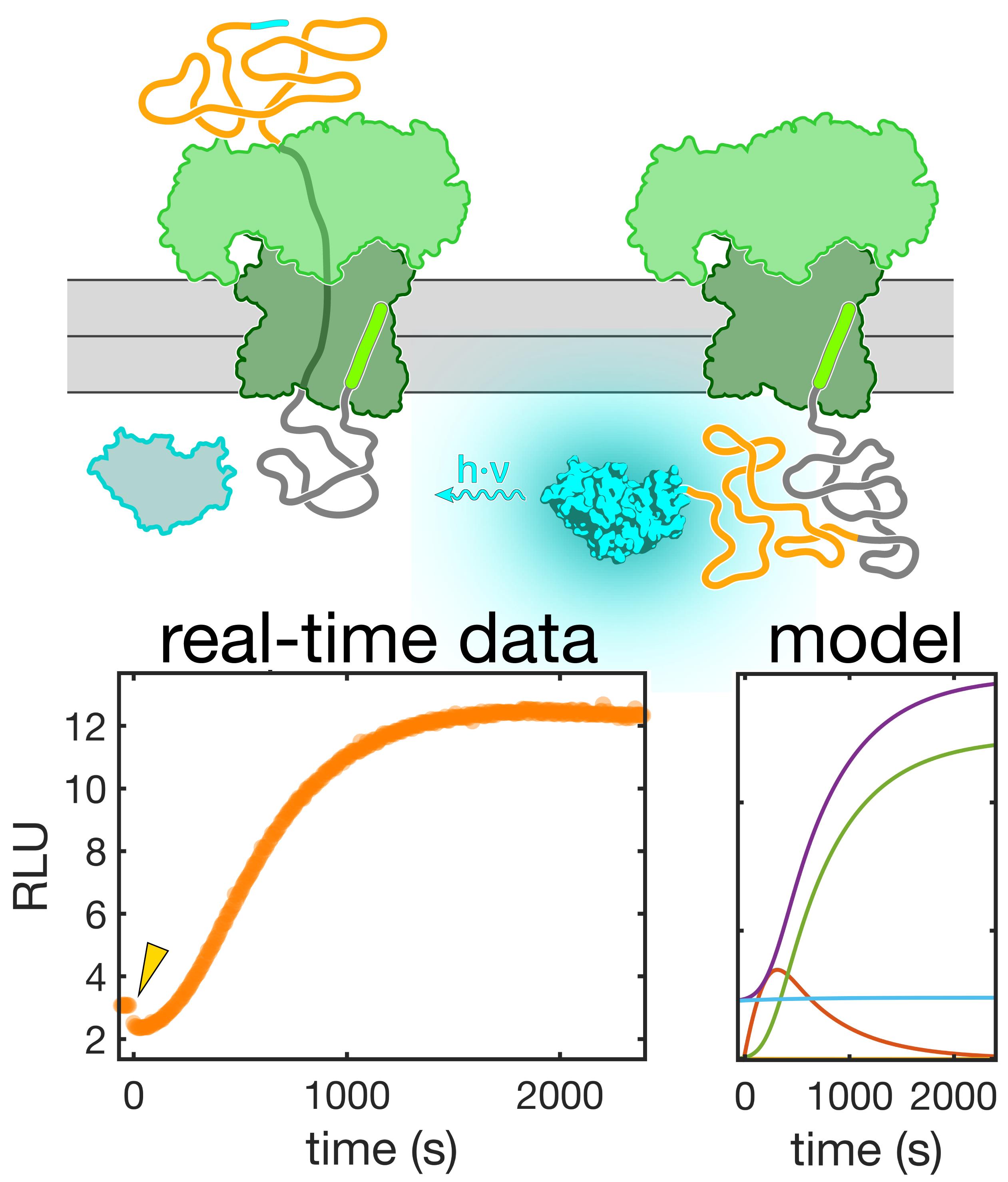
Synchronized real-time measurements, combined with a detailed kinetic model, enable a mechanistic analysis of protein transport.
Background
The Sec translocon moves proteins across lipid bilayers in all domains of life. In bacteria, a heterotrimeric transmembrane channel (SecYEG), located in the plasma membrane, associates with an ATPase, SecA, which drives substrate protein translocation. SecA utilizes chemical energy from ATP to drive the reaction forward and help to destabilize the tertiary structure of the substrate protein that otherwise interferes with translocation progress. Substrate proteins pass through the channel in an unfolded conformation.
The exact mechanisms underlying translocon function have remained unclear despite decades of study. Mechanistic studies of Sec-dependent protein translocation have traditionally been carried out using protease protection experiments (Cunningham et al., 1989). Substrate proteins are imported into vesicles or proteoliposomes containing reconstituted translocon complexes. The addition of a non-specific protease to the reaction results in proteolysis of untranslocated polypeptides, whereas proteins translocated into the interior of the proteoliposomes are protected from digestion. The protected undigested proteins are subsequently quantitated after separation by gel electrophoresis. These measurements have been instrumental in elucidating the function of the Sec translocon [see for instance (Brundage et al., 1990; Schiebel et al., 1991; van der Wolk et al., 1997; Mao et al., 2013; Bauer et al., 2014; Mao et al., 2020)]. However, the time resolution of this assay is limited because samples need to be taken and processed individually for each timepoint; thus, detailed kinetic information about the translocation process cannot be obtained.
Recently, an assay for continuously monitoring translocation has been described (Pereira et al., 2019). This assay is based on complementation of a highly active luciferase, NanoLuc, which can be asymmetrically split into a large (18 kDa) and small (1.3 kDa) fragment, termed 11S and p86, respectively (Dixon et al., 2016). Neither fragment alone exhibits significant luciferase activity. When 11S is encapsulated into SecYEG/SecA proteoliposomes, complete translocation of a substrate protein C-terminally tagged with p86 restores luciferase activity. The high affinity of the 11S/p86 complex (KD = 700 pM (Dixon et al., 2016)) and the high catalytic activity after association of the fragments permits sensitive detection of substrate proteins accumulating inside the proteoliposomes after complete translocation.
We have combined the luminescence-based real-time translocation assay (Pereira et al., 2019) with a method of generating reversibly stalled translocation intermediates (Erlandson et al., 2008) to kinetically resolve Sec-dependent protein translocation (Gupta et al., 2020). Together with a detailed kinetic model of the underlying process (Gupta et al., 2020), this approach has allowed us to dissect individual components of the process. Here, we describe the protocols for these measurements, which should be useful for obtaining mechanistic insight into the function of the Sec translocon.
The methodology described here is ideally suited for mechanistic studies of Sec translocon activity. The continuous measurement of substrate import provides time resolution that is superior to that of the traditional protease protection assays. Together with the model that we developed for analysis, the approach enables detailed kinetic dissection of the process, providing mechanistic insight into the SecA function.
Materials and Reagents
SnakeSkinTM Dialysis Tubing, 10K MWCO, 22 mm (Thermo Fisher Scientific, catalog number: 68100)
Amicon Ultra-15 Centrifugal Filter Units (Millipore Sigma, catalog number: UFC905008)
Plasmids
pSecAΔcys: expression plasmid for N-terminally His6-tagged SecA, with all cysteines replaced with serines (Gupta et al., 2020)
pSecYEG: expression plasmid for N-terminally His6-tagged SecY, SecE, and SecG; this plasmid was a kind gift from Dr. Shu-ou Shan (Caltech)
pOA-mDHFR-p86: expression plasmid for a fusion protein containing the N-terminal 178 amino acids of proOmpA, a GSGS linker, mouse DHFR, and the p86 peptide (Dixon et al., 2016)
Note: A different protein of interest can be inserted instead of mouse DHFR after the GSGS linker and before the p86 peptide. Such a construct would allow one to use this assay to study a different protein of interest.
p11S: expression plasmid for the 11S NanoLuc fragment (Dixon et al., 2016) with an N-terminal His6-SUMO tag
pGST-dark: expression plasmid for an inactive p86 “dark” peptide fused to glutathione-S-transferase (Pereira et al., 2019)
Cells
BL21 (DE3) Competent cells (Agilent Technologies, catalog number: 230130)
MM52 competent cells [prepared according to Sambrook and Russel (2001); strain from Oliver and Beckwith (1981) #1128]
LB medium (Sambrook and Russel, 2001)
LB-agar plates with Amp (Sambrook and Russel, 2001)
Super Optimal Broth (SOB) (Sambrook and Russel, 2001)
Ampicillin (Amp) (Millipore Sigma, catalog number: 171254)
Prionex (Millipore Sigma, Sigma-Aldrich, catalog number: G0411)
Nano-Glo® Live Cell Assay System (Promega, catalog number: N2011)
cOmplete Protease Inhibitor Cocktail (Millipore Sigma, Roche, catalog number: CO-RO)
n-Dodecyl β-D-maltoside (DDM) (Millipore Sigma, Sigma-Aldrich, catalog number: D4641)
1,10-phenanthroline (Ph) (Millipore Sigma, Sigma-Aldrich, catalog number: 131377)
E. coli polar lipid extract (Avanti, catalog number: 100600)
Bio-Beads SM-2 Resin (Bio-Rad, catalog number: 1523920)
Ulp1 protease (prepared in-house according to Mossessova and Lima, 2000)
Adenosine 5′-triphosphate (ATP) disodium salt hydrate (Millipore Sigma, catalog number: FLAAS)
Protein purification buffers (see Recipes)
Preparation of SecYEG/SecA vesicles with encapsulated 11S (see Recipes)
Preparation of oxidized pOA-mDHFR-p86 (see Recipes)
Real-time translocation measurement (see Recipes)
Equipment
Ultracentrifuge (Beckman Coulter, model: Optima MAX-XP)
High Performance Centrifuge (Beckman Coulter, model: Avanti J-E)
Incubator Shaker (Innova Model 44R)
GloMax® Navigator System (Promega, model: GM2000 and 2010)
Ultracentrifuge (Beckman, model: Optima XL-80k)
Type 45 Ti Fixed Angle Titanium Rotor (Beckman Coulter, catalog number: 339160)
Emulsiflex C5 (Avestin)
Sonicator (Branson)
AKTA pure FPLC system (Cytiva)
HisTrap HP (5 ml) (Cytiva, catalog number: 17524801)
HiTrap Q Sepharose Fast Flow Columns (1 ml) (Cytiva, catalog number: 17505301)
SP Sepharose Fast Flow cation exchange chromatography resin (Cytiva, catalog number: 17072901)
HisPurTM Ni-NTA Resin (Thermo Fisher Scientific, catalog number: 88221)
Extruder (Avanti, model: 610020)
Hi-Prep 16/60 Sephacryl S-200 (Cytiva, catalog number: 17116601)
Poly-Prep® Chromatography Columns (Bio-Rad, catalog number: 7311550)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Gupta, R., Toptygin, D. and Kaiser, C. M. (2021). Synchronized Real-time Measurement of Sec-mediated Protein Translocation. Bio-protocol 11(16): e4129. DOI: 10.21769/BioProtoc.4129.
分类
生物物理学 >
生物化学 > 蛋白质 > 活性
分子生物学 > 蛋白质 > 细胞间转移
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













