- EN - English
- CN - 中文
Construction of a Highly Diverse mRNA Library for in vitro Selection of Monobodies
体外选择单体的高度多样性mRNA文库的构建
发布: 2021年08月20日第11卷第16期 DOI: 10.21769/BioProtoc.4125 浏览次数: 3844
评审: Luis Alberto Sánchez VargasZHILEI CHENAnonymous reviewer(s)

相关实验方案
蛋白质pull-down免疫共沉淀实验分析病毒DNA结合蛋白质之间的直接相互作用
Ana Lechuga [...] Modesto Redrejo-Rodríguez
2018年01月05日 12405 阅读
Abstract
Recently, we developed transcription/translation coupled with the association of puromycin linker (TRAP) display as a quick in vitro selection method to obtain antibody-like proteins. For the in vitro selection, it is important to prepare mRNA libraries among which the diversity is high. Here, we describe a method for the preparation of monobody mRNA libraries with greater than 1013 theoretical diversity. First, we synthesized two long single-stranded DNAs that corresponded to fragments of monobody DNA, with random codons in the BC and FG loops. These oligonucleotides were ligated by T4 DNA ligase with the support of guide oligonucleotides containing 3′ ends that were protected by a modification. After amplifying the product DNAs by PCR, one end of each DNA fragment was digested with the type II restriction enzyme BsaI, and the resulting DNA fragments were ligated using T4 DNA ligase. After amplification of the DNA product, mRNAs were synthesized by T7 RNA polymerase. This method is simple and could be used for the preparation of mRNA libraries for various antibody-like proteins.
Graphic abstract:
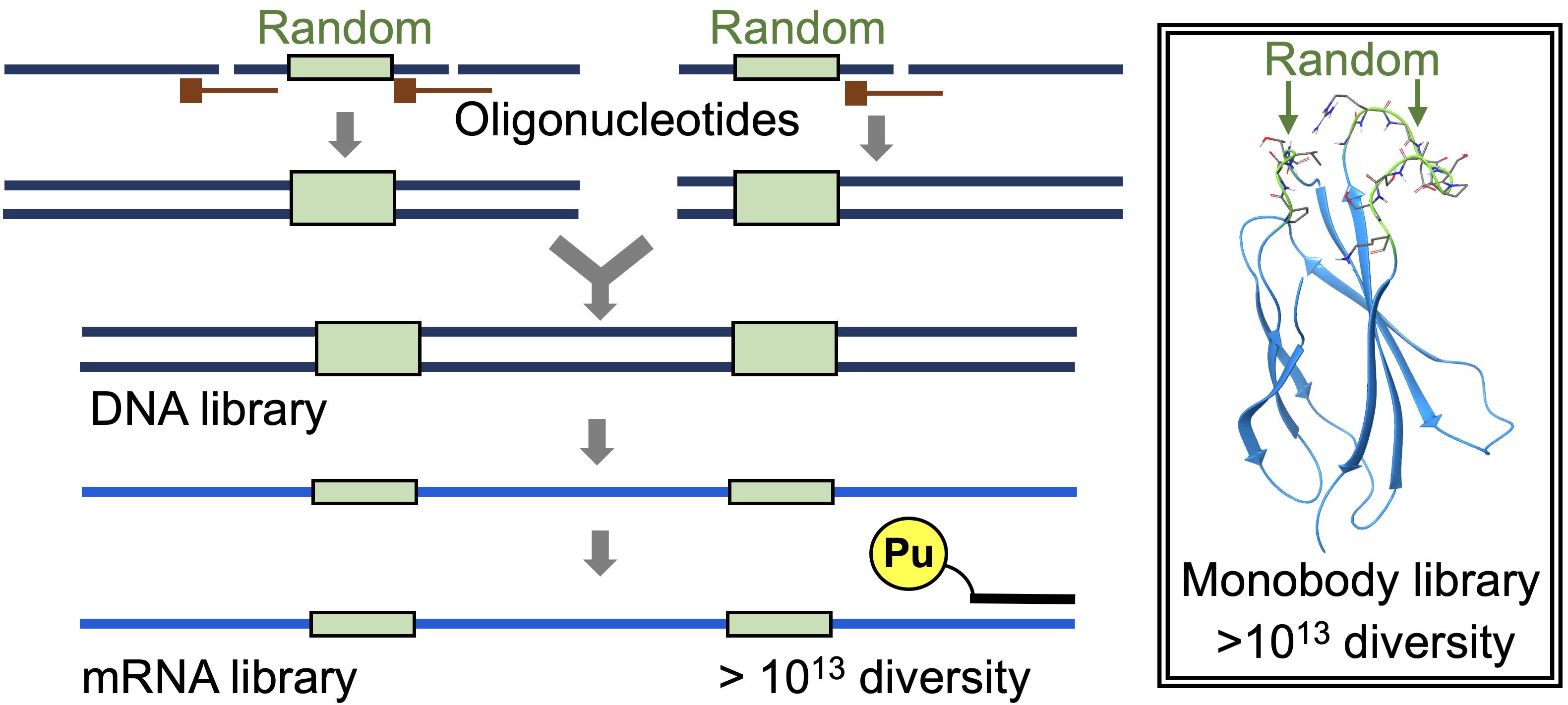
Construction of a highly diverse mRNA library.
Background
Various antibody-like proteins (ALPs) have been obtained using an in vitro selection approach and employed in the biological and medical fields (Schumacher et al., 2018; Simeon and Chen 2018). During this in vitro selection, initial library construction is an important step in obtaining high-affinity binders. For example, to develop a nanobody, the VHH domains of camel antibodies obtained after immunization were used for the construction of an initial library (Arbabi Ghahroudi et al., 1997). Alternatively, the VHH domains from naïve genes or a semi-synthetic or synthetic library could be used as the initial library (Goldman et al., 2006; Monegal et al., 2009; Yan et al., 2014).
For other non-immunoglobulin scaffolds, synthetic libraries were used exclusively as the initial library (Nord et al., 1997; Koide et al., 1998; Beste et al., 1999; Binz et al., 2003; Grabulovski et al., 2007; Olson et al., 2008). Oligonucleotides containing random codons were assembled by extension PCR and ligated to other DNA fragments for synthesis of the DNA library. As randomized codons, NNK codons (with N representing A, C, G, or T; and K representing T or G) were generally used because they cover all 20 amino acids and can reduce the redundancy of codons and the frequency of stop codons. Alternatively, direct codon synthesis using trinucleotide phosphoramidites (Virnekäs et al., 1994) was used to mimic the amino acid frequency in the CDR3 sequences of antibodies (Zemlin et al., 2003; Wojcik et al., 2010; Koide et al., 2012).
The construction of medium-sized libraries (1010 diversity) is relatively simple, whereas that of large-sized libraries (1013 to 1014 diversity) requires special care to maintain the diversity (Olson and Roberts 2007). For example, when a 50 nM DNA product is obtained after 10 cycles of PCR, the concentration of the original template can be calculated as 50 pM. If we use only 100 µl reaction mixture, the maximum diversity that can be achieved using the conditions described above is limited to 3 × 109.
Recently, by modifying the traditional mRNA display (Nemoto et al., 1997; Roberts and Szostak 1997) and the original version of the TRAP display (Ishizawa et al., 2013; Kawakami et al., 2013), we developed an improved TRAP display for the quick in vitro selection of ALPs from a large library (>1013 theoretical diversity), from which we selected high-affinity monobodies (sub-nM KD) against the SARS-CoV-2 spike protein (Kondo et al., 2020). In the present article, we describe the preparation of a monobody mRNA library with >1013 diversity (Figures 1 and 2). We first divided the monobody gene into two fragments, one containing 8 or 10 random codons in the BC loop sequence (A-fragment) and the other containing 10 or 12 random codons in the FG loop sequence (B-fragment). As a random codon, we used a codon mix with the following ratios: 20% Tyr, 10% Ser, 15% Gly, 10% Trp, and 3% each of all remaining amino acids, with the exception of Cys, which was similar to the original cocktail (30% Tyr, 15% Ser, 10% Gly, 5% Phe, 5% Trp, and 2.5% each of all remaining amino acids, with the exception of Cys) (Wojcik et al., 2010; Koide et al., 2012). To prepare a template DNA for each of the two fragments, we ligated 2 to 3 oligonucleotides assembled by guide DNAs using the T4 DNA ligase. We added a modification at the 3′ end of the guide DNAs to prevent them from becoming a primer in the following amplification step. After amplifying the product DNAs by large-scale PCR with minimum amplification cycles (15 ml, 7 cycles), these DNA fragments were digested with BsaI and ligated using T4 DNA ligase. After amplification of the DNA product by large-scale PCR (60 ml, 4 cycles), mRNAs were synthesized by T7 RNA polymerase. This procedure would be useful for the preparation of high-diversity mRNA libraries for various ALPs in the future.

Figure 1. Monobody library sequences. A. DNA sequence of the monobody library [T7 promoter, green; codon mix (20% Tyr, 10% Ser, 15% Gly, 10% Trp, and 3% each of all remaining amino acids, with the exception of Cys), yellow; an21 sequence (the 21-mer sequence for annealing to HEX-PuL), cyan]. B. Protein sequence of the monobody library (randomized residues X, yellow).
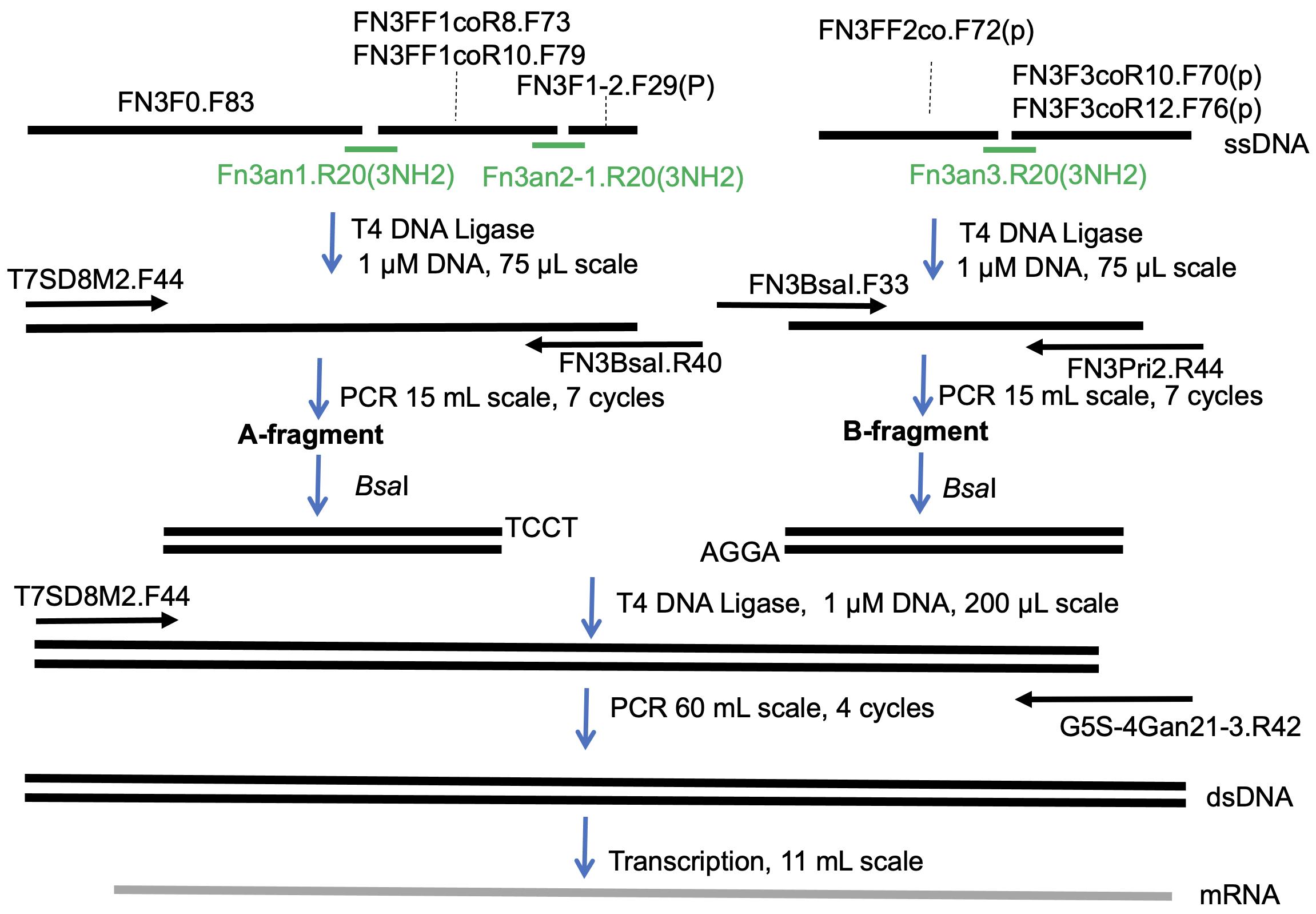
Figure 2. Scheme used for preparation of the monobody mRNA library.
Materials and Reagents
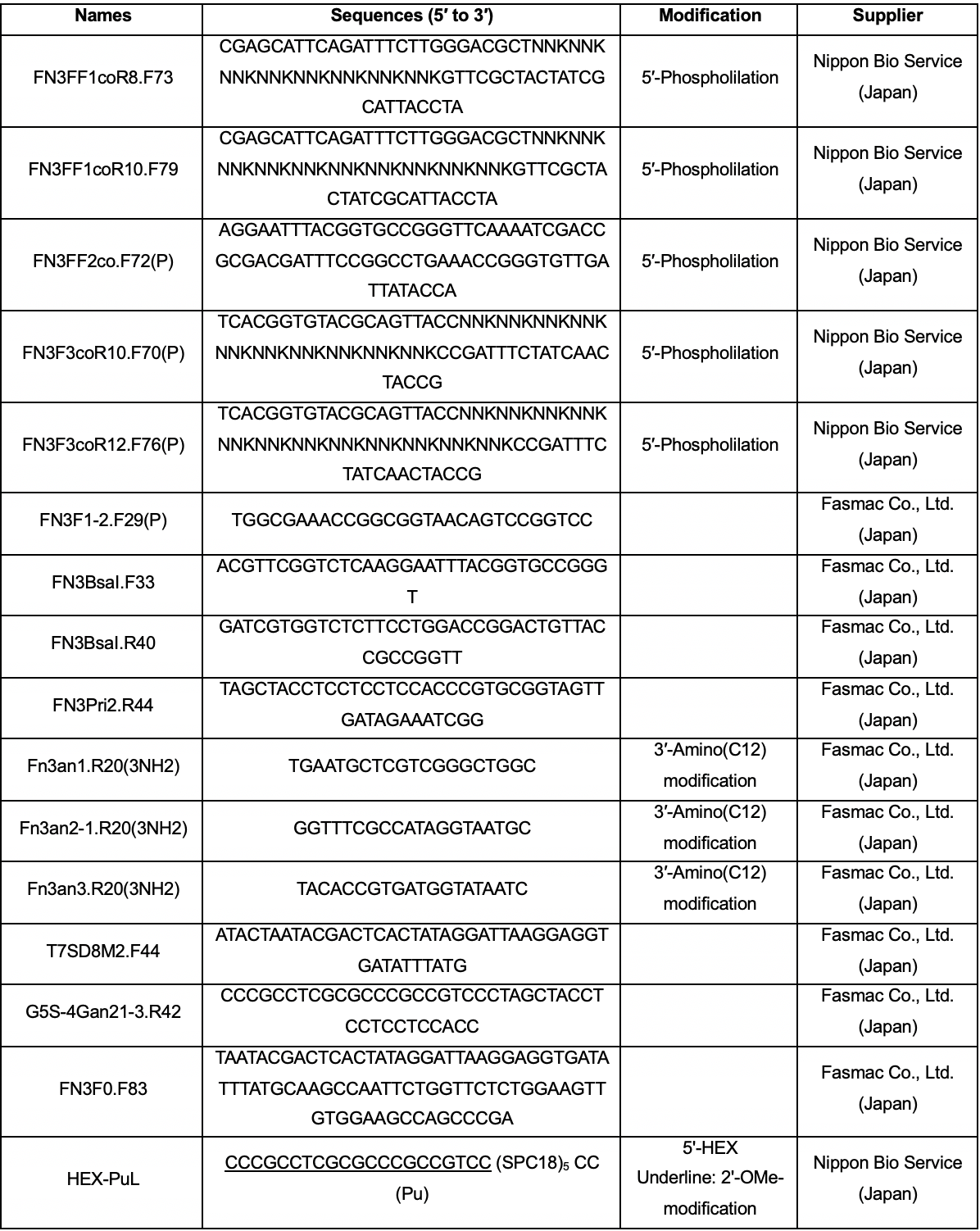
Oligonucleotides (sequences and suppliers are listed in Table 1)
T4 DNA ligase (New England Biolabs, catalog number: M0202L)
Bsa I-HF® (New England Biolabs, catalog number: R3535L)
Note: This has been replaced with a new version (Bsa I-HFv2®, catalog number: R3733L).
Pfu-S DNA polymerase prepared in-house (Wang et al., 2004; Kondo et al., 2020)
T7 RNA polymerase prepared in-house (Abramochkin and Shrader, 1995; Kondo et al., 2020)
ExcelBand 100-bp DNA Ladder (Smobio Technology, catalog number: DM2100)
0.5 mol/L-EDTA Solution (pH 8.0) (Nacalai Tesque, catalog number: 14347-21)
Formamide (Nacalai Tesque, catalog number: 16229-95)
Glycerol (Nacalai Tesque, catalog number: 17018-25)
40 (w/v) %-Acrylamide/Bis Mixed Solution (37.5:1) (Nacalai Tesque, catalog number: 06121-95)
Trimethylolaminomethane (Tris) (Nacalai Tesque, catalog number: 35406-75)
Polyethylene Glycol 6000 (PEG 6000) (FUJIFILM Wako Pure Chemicals, catalog number: 169-22945)
1,4‐Dithiothreitol (DTT) (Nacalai Tesque, catalog number: 14128-62)
Magnesium Chloride Hexahydrate (MgCl2) (Nacalai Tesque, catalog number: 20908-65)
Potassium Chloride (KCl) (Nacalai Tesque, catalog number: 28514-75
Sodium Chloride (NaCl) (Nacalai Tesque, catalog number: 31320-05)
Polyethylene Glycol Mono-p-isooctylphenyl Ether (Triton X-100) (Nacalai Tesque, catalog number: 12967-45)
Dimethyl Sulfoxide (DMSO) (Nacalai Tesque, catalog number: 13407-03)
Magnesium Sulfate Heptahydrate (Nacalai Tesque, catalog number: 06296-25)
Spermidine Trihydrochloride (Nacalai Tesque, catalog number: 32110-54)
100 mM dNTP Set (NIPPON GENE CO., LTD., Custom order)
CTP, GTP, UTP (JENA BIOSCIENCE, Custom order)
ATP (FUJIFILM Wako Pure Chemicals, catalog number: 019-09672)
Phenol:Chloroform:Isoamyl Alcohol 25:24:1 Mixed, pH 6.7 (Nacalai Tesque, catalog number: 25967-74)
Chloroform (Nacalai Tesque, catalog number: 08402-55)
2-Propanol (Nacalai Tesque, catalog number: 29113-95)
Ethanol (EtOH) (Nacalai Tesque, catalog number: 14713-95)
2× Ligation solution (see Recipes)
1× PCR solution (see Recipes)
1× RNA Pol solution (see Recipes)
DNA loading buffer (see Recipes)
Table 1. Oligonucleotide sequences and suppliers. NNK codons were prepared via trinucleotide phosphoramidite synthesis (20% Tyr, 10% Ser, 15% Gly, 10% Trp, and 3% each of all the remaining amino acids, with the exception of Cys). Abbreviations: SPC18, spacer 18; Pu, puromycin; HEX, hexachloro-fluorescein.
Equipment
T100 thermal cycler (Bio-Rad, catalog number: 1861096J1)
Gel Doc EZ (Bio-Rad, catalog number: 1708270)
Mini-PROTEAN System (Bio-Rad, catalog number: 1658000FC)
Cooling slab electrophoresis device (BIO CRAFT, catalog number: BE130G)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kondo, T., Eguchi, M., Tsuzuki, N., Murata, N., Fujino, T., Hayashi, G. and Murakami, H. (2021). Construction of a Highly Diverse mRNA Library for in vitro Selection of Monobodies. Bio-protocol 11(16): e4125. DOI: 10.21769/BioProtoc.4125.
- Kondo, T., Iwatani, Y., Matsuoka, K., Fujino, T., Umemoto, S., Yokomaku, Y., Ishizaki, K., Kito, S., Sezaki, T., Hayashi, G. and Murakami, H. (2020). Antibody-like proteins that capture and neutralize SARS-CoV-2.Sci Adv 6(42).
分类
免疫学 > 抗体分析
微生物学 > 微生物生物化学 > 蛋白质 > 免疫检测
生物化学 > 蛋白质
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link