- EN - English
- CN - 中文
Fe-NTA Microcolumn Purification of Phosphopeptides from Immunoprecipitation (IP) Eluates for Mass Spectrometry Analysis
Fe-NTA微柱纯化免疫沉淀(IP)洗脱物中磷酸肽的质谱分析
发布: 2021年08月05日第11卷第15期 DOI: 10.21769/BioProtoc.4113 浏览次数: 3849
评审: Khyati Hitesh ShahYu JinAnonymous reviewer(s)

相关实验方案

用于比较人冷冻保存 PBMC 与全血中 JAK/STAT 信号通路的双磷酸化 CyTOF 流程
Ilyssa E. Ramos [...] James M. Cherry
2025年11月20日 2351 阅读
Abstract
Protein phosphorylation is a nearly universal signaling mechanism. To date, a number of proteomics tools have been developed to analyze phosphorylation. Phosphoproteome-wide analyses using whole cell extracts suffer from incomplete coverage, often missing phosphorylation events from low-abundance proteins. In order to increase coverage of phosphorylation sites on individual proteins of interest (“phospho-mapping”), immunoprecipitation (IP) followed by phosphoenrichment is necessary. Unfortunately, most commercially available phosphoenrichment kits are not readily scalable to the low-microgram quantities of protein present in IP eluates. Here, we describe a simple method specifically optimized for the enrichment of phosphopeptides from IP samples using an Fe-NTA based method. This method can be added downstream of any standard immunoprecipitation protocol and upstream of any MS analysis pipeline. The protocol described herein is cost effective, uses commonly available laboratory reagents, and can be used to obtain deep coverage of individual protein phosphorylation patterns, supplementary to phosphoproteomics data.
Graphical abstract:
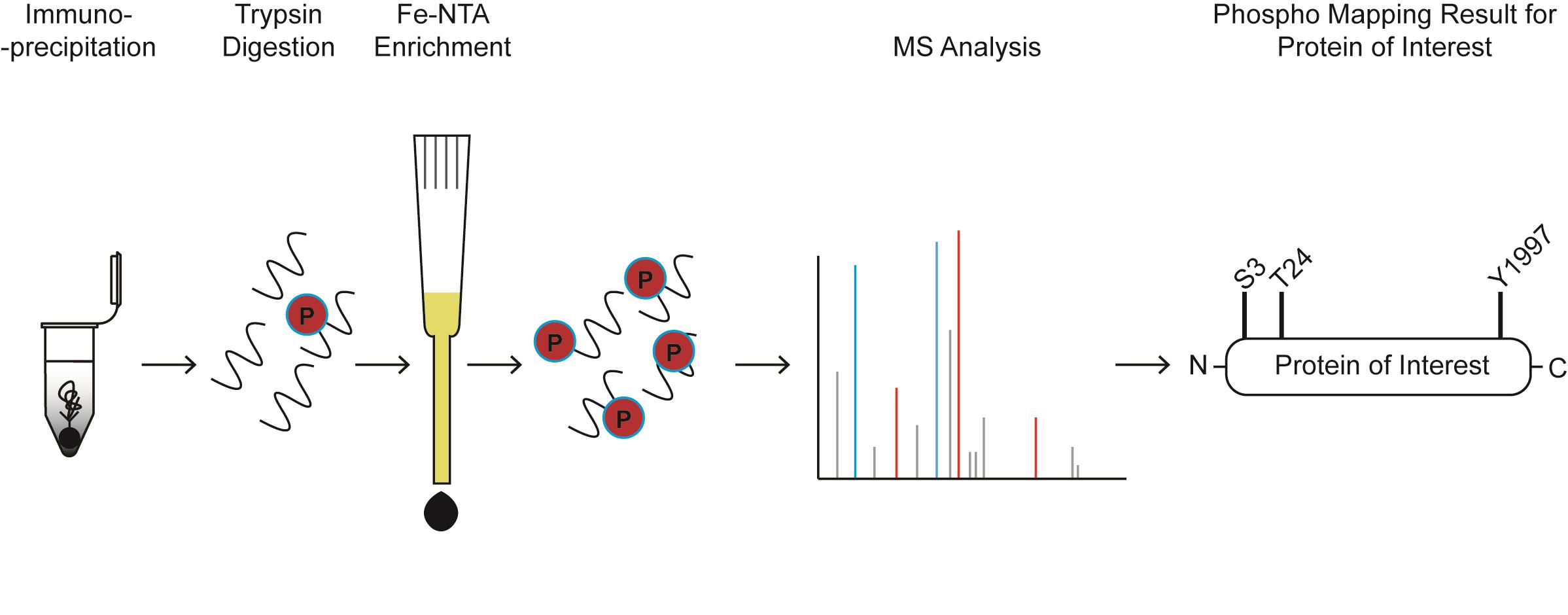
Phospho-mapping workflow for a hypothetical protein of interest
Background
Phosphorylation regulates key cellular processes such as transcription, translation, nutrient sensing, and the DNA damage response (Gingras et al., 2001; Chapman et al., 2007; Ohouo et al., 2010; Kim et al., 2011; Cussiol et al., 2015). Given the ubiquity of protein phosphorylation, its importance in normal cellular function, and its potential for dysregulation in disease, a number of proteomics methods have been developed to identify and quantitate phosphorylation events (Dephoure et al., 2013; Humphrey et al., 2018; Li et al., 2019; Faca et al., 2020). Many of these methods for phosphopeptide purification and analysis are performed on the proteome-wide scale and may suffer from relatively low coverage of individual proteins, especially those with limited abundance. High coverage of phosphorylation sites in individual proteins is often necessary to comprehensively study a given protein and to increase the chances of generating mutations of phospho-acceptor residues that elicit a phenotype. One strategy of increasing coverage of individual proteins is to first conduct an immunoprecipitation (IP) of the protein of interest using either an antibody raised against the protein itself or a common recombinant tag such as FLAG, HA, or V5. Phosphopeptides can then be isolated from the IP eluate. Optionally, differences in phosphorylation patterns between mutants or treatments can be analyzed using quantitative mass spectrometry methods such as SILAC or TMT (Ong et al., 2002; Zhang and Elias, 2017).
There are a number of commercially available kits for phosphopeptide enrichment, although these are difficult to scale down to the low-microgram quantities of protein yielded by immunopurification. There is a paucity of existing protocols for phosphopeptide enrichment following immunopurification, and those that do exist often require costly titanium dioxide (TiO2) reagents, making it difficult to scale up to multiple IPs across a panel of bait proteins (Breitkopf and Asara, 2012). Iron-based phospho-enrichment takes advantage of the affinity of negatively charged phosphate groups toward Fe3+ cations and is a cheaper alternative to TiO2; indeed, even where TiO2 is feasible, iron-based resin can be used to collect additional phospho-site data since the two methods display different phosphopeptide specificities (Bodenmiller et al., 2007).
The protocol described herein uses readily available Ni-NTA silica-based resin columns. The NTA-coupled resin is stripped of nickel, loaded with iron, and finally, the IP eluate is passed over the Fe-NTA resin to isolate phosphorylated peptides and remove unphosphorylated peptides. The procedure is performed using a home-made column (a gel-loading tip). The amount of resin generated from one Ni-NTA column is sufficient for 4-5 enrichments, making this protocol highly cost effective. Reproducibility is internally monitored via the inclusion of α/β casein phosphopeptides as controls for phosphopeptide enrichment. While this protocol has been primarily used for immunopurifications from yeast and mammalian cells, it is usable for any organism for which antibodies or recombinant tagging are available.
Materials and Reagents
C18 Sep-Pak column (Waters, catalog number: WAT043395)
Capillary tubing, 125 µm inner diameter (Polymicro, catalog number: 1068151718)
Centrifuge tubes, 50 ml (VWR, catalog number: 21008-178 or similar)
Glass fiber (Corning, catalog number: UX-34552-01)
Gel-loading pipette tips 1-200 µl (Fisher, catalog number: 02-707-138)
Low-protein binding microcentrifuge tubes, 1.5 ml (Eppendorf, catalog number: 022431081)
Microcentrifuge tubes, 1.5 ml (Eppendorf, catalog number: 0030108442)
Micro-spin columns (Thermo, catalog number: 89879)
Kimwipes (Fisher, catalog number: 06-666-1A)
Parafilm (VWR, catalog number: 52858-076)
Qiagen Ni-NTA spin columns (Qiagen, catalog number: 31014)
Razor blades (VWR, catalog number: 55411-055)
Sample vials (Sun SRI, catalog number: 501-300)
Sample vial snap caps, 11 mm (Fisher, catalog number: 14-823-379)
Acetic acid (Fisher, catalog number: A38S-500)
Acetone (Fisher, catalog number: A11-1)
Acetonitrile (Fisher, catalog number: A998-1)
Alpha casein (Sigma, catalog number: C6780-250MG)
Ammonium hydroxide solution (Sigma, catalog number: 205840010)
Angiotensin II peptide (Sigma, catalog number: A9525-5MG)
Beta casein (Sigma, catalog number: C6905-250MG)
Dithiothreitol (Sigma, catalog number: 10197777001)
Ethanol (VWR, catalog number: 89125-186)
Ethylenediamine tetraacetic acid (VWR, catalog number: MK258012)
Formic acid (Sigma, catalog number: 33015-500ML)
Iodoacetamide (Sigma, catalog number: I1149-5G)
Iron (III) chloride hexahydrate (Honeywell, catalog number: 236489-500G)
NaCl (Promega, catalog number: PRH5273)
Sodium dodecyl sulfate (Sigma, catalog number: L3771-100G)
Target polypropylene conical insert (Thermo, catalog number: C4010-629P)
Trifluoroacetic acid (Thermo, catalog number: 28904)
Tris base (Fisher, catalog number: BP152-500)
Trypsin Gold, mass spectrometry grade (Promega, catalog number: V5280)
Urea (Fisher, catalog number: U15-500)
Urea/Tris solution (see Recipes)
α/β casein peptide digest (see Recipes)
0.1 P solution (see Recipes)
Trypsin Gold solution (see Recipes)
C18 Buffer E (see Recipes)
C18 Buffer W1 (see Recipes)
C18 Buffer W2 (see Recipes)
C18 Buffer W3 (see Recipes)
FeCl3 solution (see Recipes)
IMAC elution solution (see Recipes)
IMAC stripping solution (see Recipes)
IMAC wash I (see Recipes)
Iodoacetamide (IAc) solution (see Recipes)
IP elution buffer (see Recipes)
PPT solution (see Recipes)
Equipment
1 ml BD syringes with slip-tip (BD, catalog number: 309659)
2× water bath or heat block (Thermo, model: TSGP02 or similar)
Tweezers (VWR, catalog number: 89259-984)
Desktop centrifuge (Beckman, model: A99469 or similar)
Desktop microcentrifuge (Beckman, model: A46471 or similar)
Micropipettes (Gilson, catalog number: F167380 or similar kit with P2, P20, P200, P1000 types)
Note: One water bath/heat block should be set to 42°C and the other to 65°C.Orbitrap mass spectrometer; for example, Q-Exactive HF (Thermo, IQLAAEGAAPFALGMBFZ)
Scissors (VWR, catalog number: 82027-596 or similar)
UHPLC system coupled to a mass spectrometer; for example, UltiMate 3000 (Thermo, model: IQLAAAGABHFAPBMBEX)
Vacuum concentrator centrifuge (Thermo, model: SPD131DDA)
Vacuum concentrator filter (Thermo, model: VPOF110)
Vacuum concentrator pump (Thermo, model: VLP120)
Vacuum concentrator vapor trap (Thermo, model: RVT5105)
Note: Items 9-12 are individual parts of what is referred to as a “speedvac” in the later steps of the protocol.
Software
XCalibur Qual Browser (Thermo Scientific, https://www.thermofisher.com/order/catalog/product/OPTON-30965?SID=s rch-srp-OPTON-30965#/OPTON-30965?SID=srch-srp-OPTON-30965)
Comet (http://comet-ms.sourceforge.net/)
Note: Any other commonly available mass spectrometry analysis software can be used.
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sanford, E. J. and Smolka, M. B. (2021). Fe-NTA Microcolumn Purification of Phosphopeptides from Immunoprecipitation (IP) Eluates for Mass Spectrometry Analysis. Bio-protocol 11(15): e4113. DOI: 10.21769/BioProtoc.4113.
分类
分子生物学 > 蛋白质 > 磷酸化
发育生物学 > 细胞信号传导
癌症生物学 > 基因组不稳定性及突变
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










