- EN - English
- CN - 中文
Electron Tomography to Study the Three-dimensional Structure of the Reovirus Egress Pathway in Mammalian Cells
电子断层扫描研究哺乳动物细胞呼肠孤病毒出口途径的三维结构
发布: 2021年07月05日第11卷第13期 DOI: 10.21769/BioProtoc.4080 浏览次数: 4400
评审: Juan Facundo Rodriguez AyalaKristin L. ShinglerAnca Flavia Savulescu
Abstract
Mammalian orthoreoviruses (reoviruses) are nonenveloped, double-stranded RNA viruses that replicate and assemble in cytoplasmic membranous organelles called viral inclusions (VIs). To define the cellular compartments involved in nonlytic reovirus egress, we imaged viral egress in infected, nonpolarized human brain microvascular endothelial cells (HBMECs). Electron and confocal microscopy showed that reovirus mature virions are recruited from VIs to modified lysosomes termed sorting organelles (SOs). Later in infection, membranous carriers (MCs) emerge from SOs and transport new virions to the plasma membrane for nonlytic egress. Transmission electron microscopy (TEM) combined with electron tomography (ET) and three-dimensional (3D) reconstruction revealed that these compartments are connected and form the exit pathway. Connections are established by channels through which mature virions are transported from VIs to MCs. In the last step, MCs travel across the cytoplasm and fuse with the plasma membrane, which facilitates reovirus egress. This bio-protocol describes the combination of imaging approaches (TEM, ET, and 3D reconstruction) to analyze reovirus egress zones. The spatial information present in the 3D reconstructions, along with the higher resolution relative to 2D projections, allowed us to identify components of a new nonlytic viral egress pathway.
Keywords: Reovirus (呼肠孤病毒)Background
Viruses recruit and transform cellular compartments to build membranous viral factories or inclusions, where genome replication and particle assembly take place (Miller and Krijnse-Locker, 2008; Sachse et al., 2019). Following genome packaging and assembly, progeny virions leave the host cell. Both enveloped and nonenveloped viruses can induce cell death by lysis, resulting in the release of viral particles. However, viral egress pathways can also be mediated by intracellular organelles that transport new virions to the plasma membrane, allowing them to exit cells without lysis (Roth et al., 2020). Therefore, many viruses use cellular membranous organelles for replication and assembly, as well as for egress and cell-to-cell transmission (Altan-Bonnet, 2017; Bird and Kirkegaard, 2015).
Reoviruses belong to the Reoviridae family, which includes several pathogens of plants, animals, and humans. Reoviruses are nonenveloped, double-stranded RNA viruses that assemble membranous cytoplasmic structures termed VIs. Early in infection, reovirus nonstructural proteins σNS and μNS induce the remodeling of ER membranes to form VIs (Tenorio et al., 2018). Viral genome replication, secondary rounds of transcription, and capsid assembly occur in VIs and, as a result, mature virions as well as empty capsids gather in this network of membranes. Reovirus uses different types of egress mechanisms depending on the cell type and culture conditions. In some types of cultured cells, reovirus infection leads to NF-κB activation, inducing apoptotic signaling and eventually lytic cell death (Danthi et al., 2013); however, reovirus can undergo nonlytic egress in other cell types such as HBMECs (Lai et al., 2013). In nonpolarized HBMECs, progeny virions exit from discrete areas at the basal surface. The detailed study of these regions is difficult because of their low frequency and location at the cell base. Due to these constraints, late steps of reovirus infection, including nonlytic egress, are not well understood.
The use of several imaging techniques in combination allowed us to discover that the reovirus egress pathway is composed of two different membranous elements called SOs and MCs. SOs are modified lysosomes that are recruited to the periphery of VIs during late phases of infection. Only mature genome-containing virions are collected in SOs, whereas empty capsids are absent. The smaller MCs are formed by budding from SOs and transport viral progeny to the plasma membrane (Fernandez de Castro et al., 2020). ET and 3D reconstruction were essential in defining the spatial relationships of the different compartments and elucidating the intracellular reovirus egress pathway (a summary of the protocol is shown in Figure 1).
ET can be used to visualize the structure of viruses, cells, and their constituents in three dimensions. The chosen approach defines the volume that can be analyzed as well as the possible resolution that can be obtained. In TEM-ET, micrographs of the sample are obtained from various orientations by tilting the specimen, and the resulting micrographs are computationally combined to form a 3D volume (Ercius et al., 2015, Hoppe 1974). Developments in hardware and software during recent years have allowed automatization of the image acquisition process. These innovations enable the production of tomograms, which allows appreciation of cellular organelle ultrastructure and inter-organelle connections in unprecedented detail in 3D. The 3D volumes are useful for studies to determine how cellular compartments interact since they display the spatial organization of structures within the volume, whereas a single 2D image provides only a projection of this information. Interestingly, 3D-ET data showed connections between VIs, SOs, and MCs, the three main components of the reovirus egress pathway. Furthermore, we were able to determine the size of the channels that connect the egress compartments (around 90-100 nm in diameter), which could not be established by analysis of ultra-thin sections (50-70 nm). We were also able to observe that viral egress from MCs to the extracellular medium is mediated by membrane fusion events at the plasma membrane. These events were visualized using double-tilt ET, which orients the specimen around two orthogonal axes, resulting in two independent tomograms computed from each tilt series. The two tomograms were aligned with each other and combined to obtain a single tomogram. As a result, a dual-axis tomogram shows better resolution at any orientation in the plane of the specimen than a single-axis series, in which the specimen is tilted about only one axis (Mastronarde 1997).
In this Bio-protocol, we describe the use of 3D-ET to study the reovirus nonlytic egress pathway and define the morphology and connections between the cellular organelles that compose the exit route. This methodology provides great potential to characterize many other cellular pathways in which different organelles are involved and connected as well as the complexity of pathogen-cell interactions.
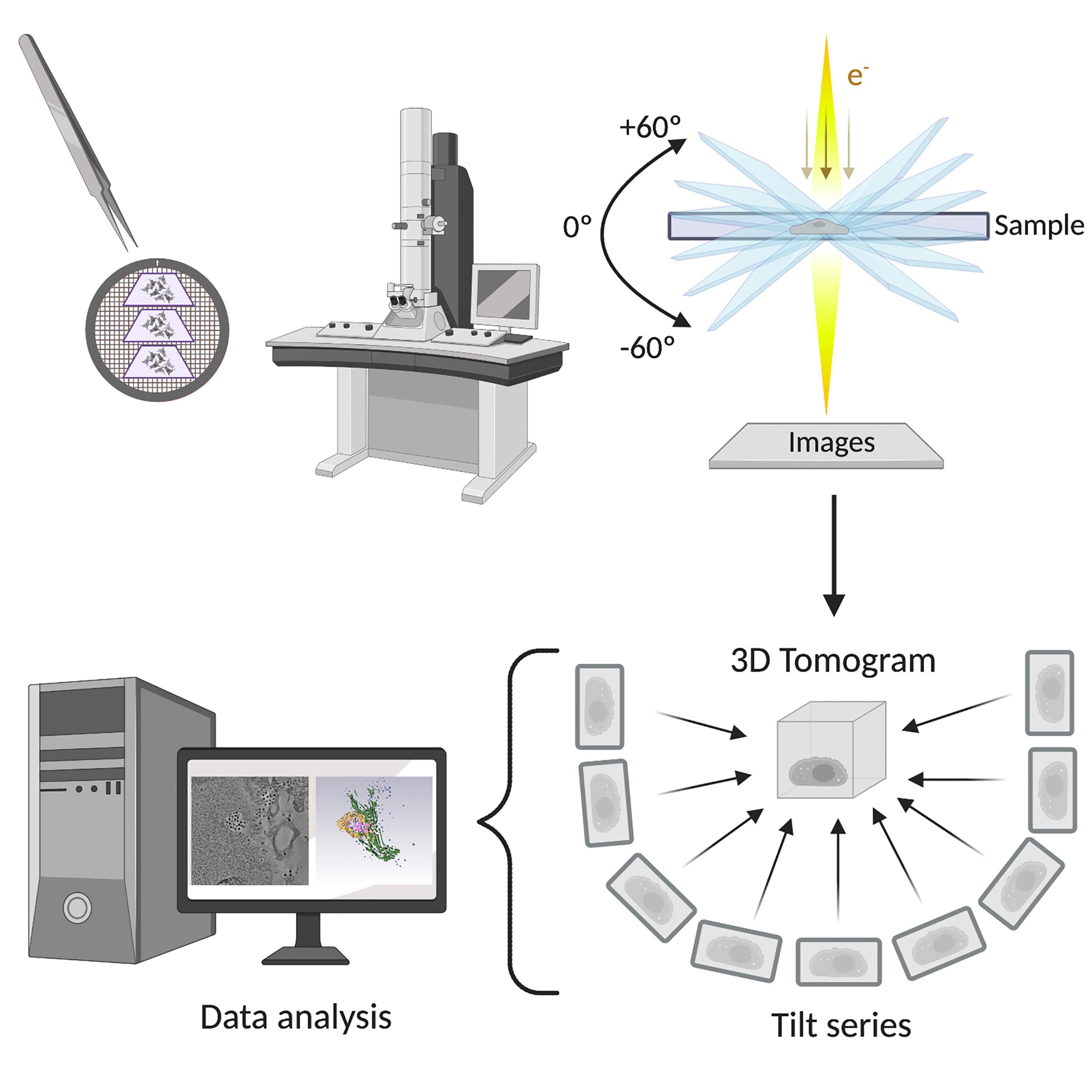
Figure 1. Electron tomography of the reovirus egress pathway workflow. Sections of reovirus-infected cells were imaged by transmission electron microscopy to discern reovirus exit zones. The structures of interest were captured, and tilt series were obtained. After image processing and reconstruction, the 3D tomogram can be analyzed. Created with BioRender.com. © 2020 Fernández de Castro et al., originally published in J Cell Biol https://doi.org/10.1083/jcb.201910131.
Materials and Reagents
Human brain microvascular endothelial cells (HBMECs) (Stins et al., 1997; Stins et al., 2001)
Reovirus strain T1L M1-P208S was recovered from plasmids by reverse genetics as described previously (Kobayashi et al., 2007 and 2009). This strain was engineered by the Terence Dermody Lab.
Roswell Park Memorial Institute (RPMI)-1640 Medium (Merck, catalog number: R8758)
Trypsin-ethylenediaminetetraacetic acid (EDTA) solution 10× (Merck, catalog number: 9002-07-7)
Fetal bovine serum (FBS) (BI Biological Industries, catalog number: 04-007-1A)
Corning® Nu-SerumTM IV Growth Medium Supplement (Corning®, Life Sciences, catalog number: 55004)
Minimum Essential Medium (MEM) vitamin solution 100× (ThermoFisher Scientific, GibcoTM, catalog number: 11120052)
Sodium pyruvate solution (Merck, Sigma-Aldrich, catalog number: 113-24-6)
MEM non-essential amino acid Solution 100× (Merck, catalog number: M7145)
L-glutamine (Merck, catalog number: 56-85-9)
Penicillin-streptomycin (Merck, Sigma-Aldrich, catalog number: P4333)
Amphotericin B solution (Merck, Sigma-Aldrich, catalog number: 1397-89-3)
4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl) piperazine-N’-(2-ethanesulfonic acid) HEPES (Merck, Sigma-Aldrich, catalog number: 7365-45-9)
Distilled 50% glutaraldehyde (TAAB Laboratories, catalog number: G015)
Sapphire disks Ø1.4 mm (Martin Wohlwend, catalog number: 454)
NuncTM cell culture-treated multidishes (ThermoFisher Scientific, catalog number: 140675)
Bovine serum albumin (BSA) (Merck, Sigma-Aldrich, catalog number: 9048-46-8)
Phosphate-buffered saline (PBS) (Merck, Sigma-Aldrich, catalog number: P3813)
Ultrapure Milli-Q water
Dumont Tweezers type 4 (TAAB Laboratories, catalog number: T052)
Dumont Tweezers type 7 (TAAB Laboratories, catalog number: T050)
Osmium Tetroxide Electron Microscopy (EM) solution (TAAB Laboratories, catalog number: O015)
Uranyl acetate (Electron Microscopy Sciences, catalog number: 22400)
Methanol dried (Merck, catalog number: 67-56-1)
Acetone dried (Merck, catalog number: 1-00299-0500)
812 resin (TAAB Laboratories, catalog number: T026)
Dodecenyl succinic anhydride (DDSA) (TAAB Laboratories, catalog number: D027)
Methyl nadic anhydride (MNA) (TAAB Laboratories, catalog number: MG12)
Benzyldimethylamine (BDMA) (TAAB Laboratories, catalog number: B022)
BEEM® embedding capsule 00 (TED PELLA, INC., catalog number: 130)
GEM® stainless-steel blade (TED PELLA, INC., catalog number: 62-0179)
Perfect loop (Electron Microscopy Sciences, catalog number: 70945)
Ultra-diamond knife 45° dry (DIATOME, catalog number: DU4530)
QUANTIFOIL® R 3.5/1 Holey Carbon Film Grids Cu 300 mesh (QUANTIFOIL®, catalog number: Q11394)
Protein A Gold 10 nm (PAG10). The source is the Cell Microscopy Core (Department of Cell Biology, University Medical Center Utrecht)
Ethanol absolute (100% ethanol) (Merck, catalog number: 64-17-5)
Liquid nitrogen
Equipment
Water purification system (Merck, Milli-Q®, model: Advantage A10)
pH meter Basic 20 (HACH LANGE SPAIN, Crison, model: Basic 20)
Carbon coating system (Leica Microsystems, model: Leica EM MED020)
Cell incubator (BINDER, model: CB 170)
High-pressure vitrification system (Leica Microsystems, model: Leica EM PACT2)
Automatic cryosubstitution system (Leica Microsystems, model: Leica EM AFS2)
Fume hood (Flow-Tronic)
Ultramicrotome (Leica Microsystems, model: Leica EM UC6)
Field Electron and Ion Company (FEI) Tecnai electron microscope (FEI, model: G2 F20 (200 kV))
Charge-couple device (CCD) camera (FEI, model: Eagle 4k×4k)
Software
Tomography 3 (https://www.thermofisher.com/) (EM software, ThermoFisher Scientific). This software was used for Tecnai imaging and tilt series acquisitions.
IMOD (Kremer et al., 1996) (https://bio3d.colorado.edu/imod/) was used for the alignment of the raw tilt series and tomogram generation.
Amira (https://www.thermofisher.com/) (2D-5D visualization and analysis software, ThermoFisher Scientific). Amira was used for tomogram segmentation, reconstruction, and visualization.
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Fernandez de Castro, I., Fernández, J. J., Dermody, T. S. and Risco, C. (2021). Electron Tomography to Study the Three-dimensional Structure of the Reovirus Egress Pathway in Mammalian Cells. Bio-protocol 11(13): e4080. DOI: 10.21769/BioProtoc.4080.
- Fernandez de Castro, I., Tenorio, R., Ortega-Gonzalez, P., Knowlton, J. J., Zamora, P. F., Lee, C. H., Fernandez, J. J., Dermody, T. S. and Risco, C. (2020). A modified lysosomal organelle mediates nonlytic egress of reovirus. J Cell Biol 219(7).
分类
细胞生物学 > 细胞结构 > 细胞器
生物物理学 > 显微技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link