- EN - English
- CN - 中文
Single-molecule Fluorescence Technique to Monitor the Co-transcriptional Formation of G-quadruplex and R-loop Structures
单分子荧光技术监测G-四链体和R-环结构的共转录形成
发布: 2021年07月05日第11卷第13期 DOI: 10.21769/BioProtoc.4069 浏览次数: 4348
评审: ilgen MenderChristina Yan Ru TanAnonymous reviewer(s)
Abstract
G-quadruplexes (GQ) and R-loops are non-canonical nucleic acid structures related to gene regulation and genome instability that can be formed during transcription; however, their formation mechanisms remain elusive. To address this question, we developed a single-molecule fluorescence technique to monitor the formation of G-quadruplex and R-loop structures during transcription. Using this technique, we found that R-loop formation precedes GQ formation and that there exists a positive feedback loop between G-quadruplex and R-loop formation.
Keywords: Single-molecule fluorescence (单分子荧光)Background
G-quadruplexes (GQ) consist of stacked G-tetrads that are formed from four Hoogsteen base-paired guanines (Gellert et al., 1962). GQ formed at certain hotspots in the genome (Biffi et al., 2013; Marsico et al., 2019) play regulatory roles (Siddiqui-Jain et al., 2002; Falabella et al., 2019) and are related to certain diseases (Biffi et al., 2014; De Magis et al., 2019). R-loops are three-stranded nucleic acid structures composed of a double-stranded DNA-RNA hybrid and a single-stranded DNA. R-loops are also related to gene regulation and genome instability (Aguilera and García-Muse 2012; Santos-Pereira and Aguilera 2015). Interestingly, GQ and R-loops are formed during transcription and often coexist (Duquette et al., 2004).
The biological roles of GQ and R-loops and their formation mechanisms have been extensively studied over the last few decades. GQ and R-loop formation has previously been detected using bisulfite sequencing (Roy and Lieber 2009) or sequencing after immunoprecipitation (Chan et al., 2014; Hansel-Hertsch et al., 2018). Fluorescent ligands that bind specifically to GQ have also been used to observe GQ formation (Kreig et al., 2015). These techniques have provided understanding of the hotspots where GQ and R-loops are often formed (Sanz et al., 2016), in addition to the enzymes that regulate the formation of GQ and R-loops (Masse and Drolet 1999); however, their exact formation mechanisms remain to be elucidated.
Here, we report single-molecule fluorescence techniques that monitor the cotranscriptional formation of GQ and R-loops. To detect GQ formation, we used FRET (Fluorescence Resonance Energy Transfer), and to detect R-loop formation, we used the fluorescently labeled S9.6 antibody (Figure 1). We monitored GQ and R-loop formation during transcription and found that R-loop formation precedes GQ formation and there exists a positive feedback loop between GQ and R-loop formation.
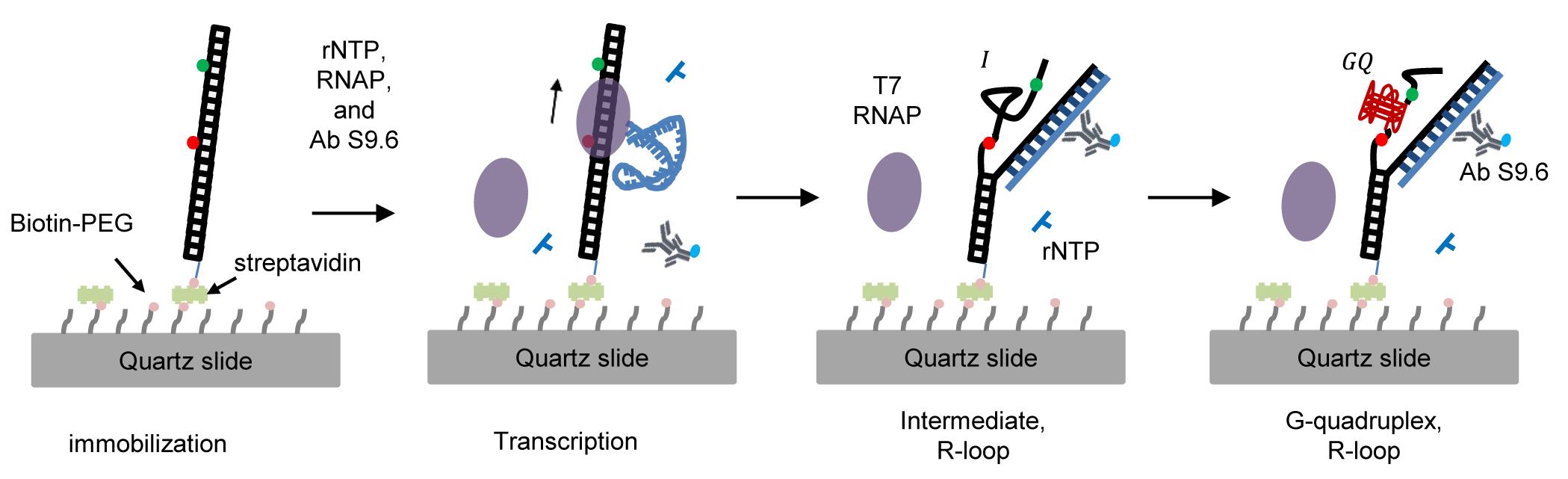
Figure 1. Single-molecule fluorescence technique to monitor the cotranscriptional formation of GQ coupled with R-loops. GQ and R-loop formation is monitored by high FRET efficiency and labeled S9.6 antibody, respectively.
Materials and Reagents
Double-sided tape (3M, catalog number: 137-ROK)
Polyethylene tubing (BD Intramedic, Fisher Scientific, catalog number: 427411)
Quartz microscope slide (H. FINKENBEINER Inc., USA, catalog number: custom-order, size : 1’’ × 3’’ × 1mm)
Micro cover glass (VWR, USA, catalog number: 48393230)
DNA oligos (Figure 2)
Non-template strand (1/2; purchased from IDT):
ATCAGGTCTAATACGACTCACTATAGGAAGAGAAAGT/iCy5/TTCTGGGAGGG
Non-template strand (2/2; purchased from IDT):
/5Phos/AGGGAGGGTGTA/iCy3/CTGATGCGTTCCACTCGC
Template strand (1/1; purchased from IDT):
GCGAGTGGAACGCATCAGTACACCCTCCCTCCCTCCCAGAAACTTTCTCTTCCTATAGTGAGTCGTATTAGACCTGAT/biotin/
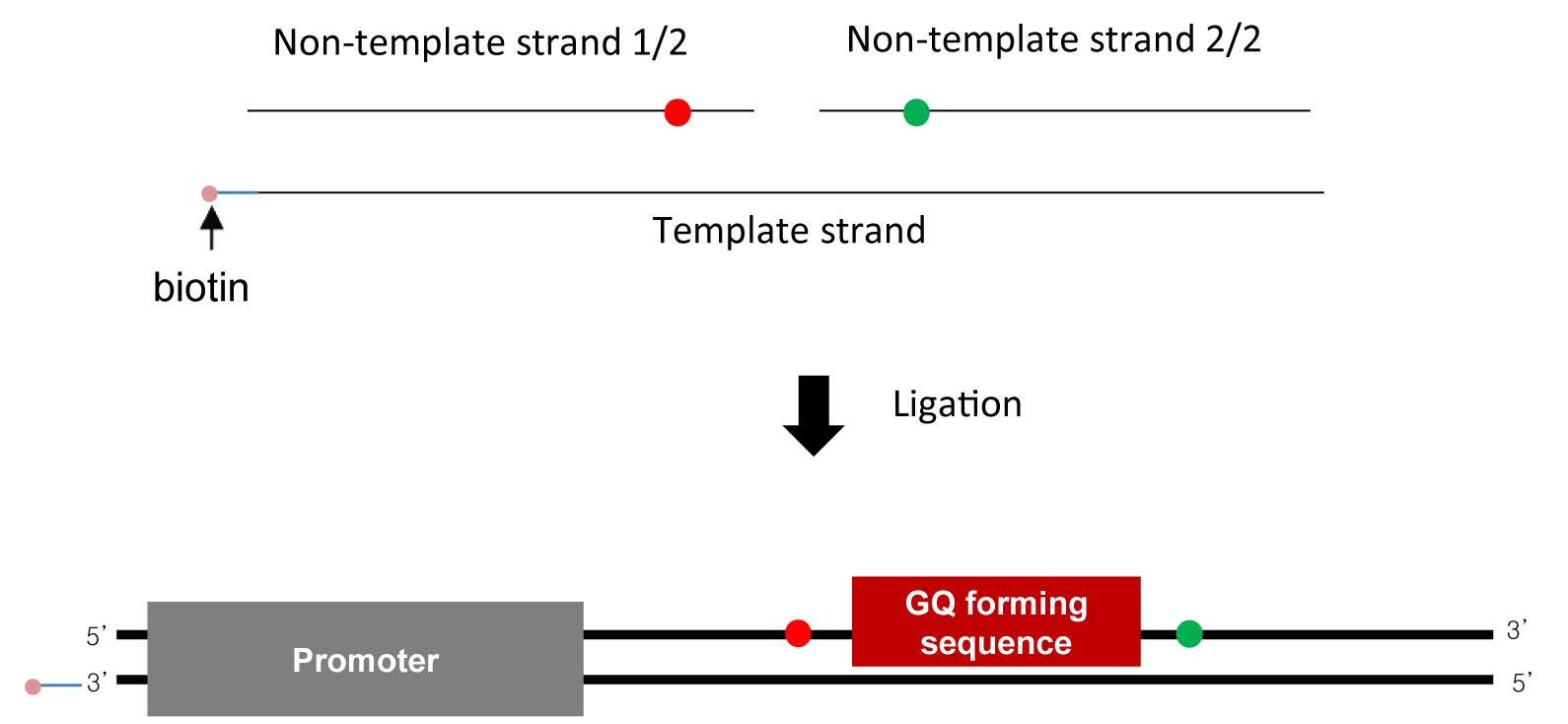
Figure 2. Sample design. DNA substrate is made using annealing and ligation (top). DNA substrate contains a T7 promoter and a GQ-forming sequence. Biotin (pink) is labeled at the upstream end for surface immobilization, and Cy3 (green) and Cy5 (red) are labeled as a FRET pair to detect GQ formation.Distilled water
Liquid nitrogen
Acrylamide:bis=29:1 (30%) (Biosesang, catalog number: A2003-1)
10× TBE (Biosesang, catalog number: TR2004-100-00)
N, N, N’, N’-tetra-methylethylenediamine (TEMED) (Bio-Rad, catalog number: 161-0801)
Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: 1001799334)
Formamide (Sigma-Aldrich, catalog number: 01246)
UREA (Bio-Rad, catalog number: 161-0731)
Ethanol (Carlo Ebra Reagents, catalog number: 528167)
Alexa Fluor 488 C5-maleimide (Thermo Fisher, Invitrogen, catalog number: A10254)
NAP-5-column (GE Healthcare, catalog number: 17-0853-02)
Primary antibody: Anti-DNA-RNA hybrid (S9.6) antibody (Kerafast, catalog number: ENH001)
Note: We divide the stock solution of S9.6 antibody into aliquots, freeze each tube in liquid nitrogen, and store them at -20°C (short term) or -80°C (long term).
Secondary antibody: Anti-Mouse IgG (H+L) (Jackson ImmunoResearch, catalog number: 715-005-151)
Sulfuric acid (Daejung, catalog number: 7664-93-9)
Hydrogen peroxide solution (30%) (DaeJung, catalog number: 7722-84-1)
Biotin-PEG-SC (Laysan Bio Inc., catalog number: 141-63)
mPEG-SVA (Laysan Bio Inc., catalog number: 149-75)
Epoxy glue (LOCTITE, catalog number: 326795)
T4 DNA ligase (New England Bio Labs, catalog number: M0202M)
10× T4 DNA ligase buffer (New England Bio Labs, catalog number: B0202S)
T7 RNA polymerase (New England Bio Labs, catalog number: M0251S)
Trolox (Sigma-Aldrich, catalog number: 238813)
PCD (Oriental Yeast Co., catalog number: 46852004)
PCA (Sigma-Aldrich, catalog number: 37580)
Spermidine (Sgima-Aldrich, catalog number: 85558)
Note: Only a small amount to be used within a week is diluted and stored in a drawer from which light is completely blocked. After using the stock solution, the air in the container should be replaced with nitrogen gas and stored at room temperature.
rNTP set (GE Healthcare, catalog number: 27202501)
1,4-dithiothreitol (Merck, catalog number: 222-468-7)
Note: DTT is known to be easily oxidized in water over time. To prevent degradation, divide 1 M DTT into aliquots and store them at -20°C after dissolving. Moreover, it is recommended not to use DTT solution several months after making it even if it has been stored at -20°C.
Acetic acid (J.T.Baker, Fisher Scientific, catalog number: 14-650-388)
Sodium bicarbonate (Fisher Scientific, catalog number: S233-500)
Sodium borate (Fisher Scientific, catalog number: S248-500)
Aminopropylsilane (UCT SPERCIALTIES, LLC, catalog number: A0700)
Fluorescent beads (FluoSpheres, Invitrogen, catalog number: 1890851)
10× TBE buffer (Biosesang, catalog number: TR2004-100-00)
PCR purification kit (QIAGEN, catalog number: 28106)
Trolox solution (see Recipes)
PCA solution (see Recipes)
T50 buffer (see Recipes)
Denaturing gel solution (see Recipes)
Piranha solution (see Recipes)
Silanization solution (see Recipes)
PEGylation solution (see Recipes)
Imaging buffer (see Recipes)
4× elongation buffer (see Recipes)
Equipment
Home-built TIRF microscope setup
Note: The components used in the home-built total internal reflection fluorescence (TIRF) microscope system are as follows: EMCCD, microscope, lasers (473-nm, 532-nm, 633-nm), shutter controller, prism, dichroic mirrors, piezo stage, and microscope temperature control system. A detailed experimental setup can be found in previously published papers (Roy et al., 2008; Lee et al., 2010).
Microscope temperature control system (Live Cell Instrument, model: CU-109)
Incubator (FINE PCR, model: ALB 6400)
Thermal cycler (Bio-Rad, model: C1000)
Vertical electrophoresis cell (Mini-PROTEAN) (Bio-Rad, model: 1658000 FC)
Gel documentation system (SYNGENE, model: G:box Chemi XT4 system)
Ultrasonic cleaner (used as a water bath) (Branson, model: 3510E-DTH)
Note: The ultrasonic cleaner can serve as a water bath in heating mode. We set the temperature to 69°C, but the actual temperature was roughly 55°C when running the denaturing gel.
Razor (Dorco, South Korea)
Diamond solid thin drill (OD: 0.75 mm) (UKAM Industrial Superhard Tools, catalog number: 2040075)
Glass jar (for Piranha)
Water purification system (Millipore, model: ZRXQ-003-EU)
Syringe pump (HARVARD APPARATUS, model: PHD2000)
Spectrophotometer (Thermo Scientific, model: NANODROP 2000)
Software
Labview (version: 2015) (National Instruments, USA, https://www.ni.com/)
IDL (version: 7.0) (David Stern & ITT Visual Information Solutions, USA, https://www.l3harrisgeospatial.com/Software-Technology/IDL)
Matlab (version: R2015a) (MathWorks, USA, https://www.mathworks.com)
Origin (version: 8.5) (Electronic Arts, USA, https://www.origin.com)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Lim, G. and Hohng, S. (2021). Single-molecule Fluorescence Technique to Monitor the Co-transcriptional Formation of G-quadruplex and R-loop Structures. Bio-protocol 11(13): e4069. DOI: 10.21769/BioProtoc.4069.
分类
生物物理学 > 显微技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










