- EN - English
- CN - 中文
Transcriptional Run-on: Measuring Nascent Transcription at Specific Genomic Sites in Yeast
连缀转录:测定酵母特定基因组位点的新生转录
发布: 2021年06月20日第11卷第12期 DOI: 10.21769/BioProtoc.4064 浏览次数: 3137
评审: Julie WeidnerLaia ArmengotAnonymous reviewer(s)
Abstract
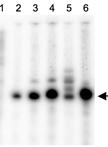
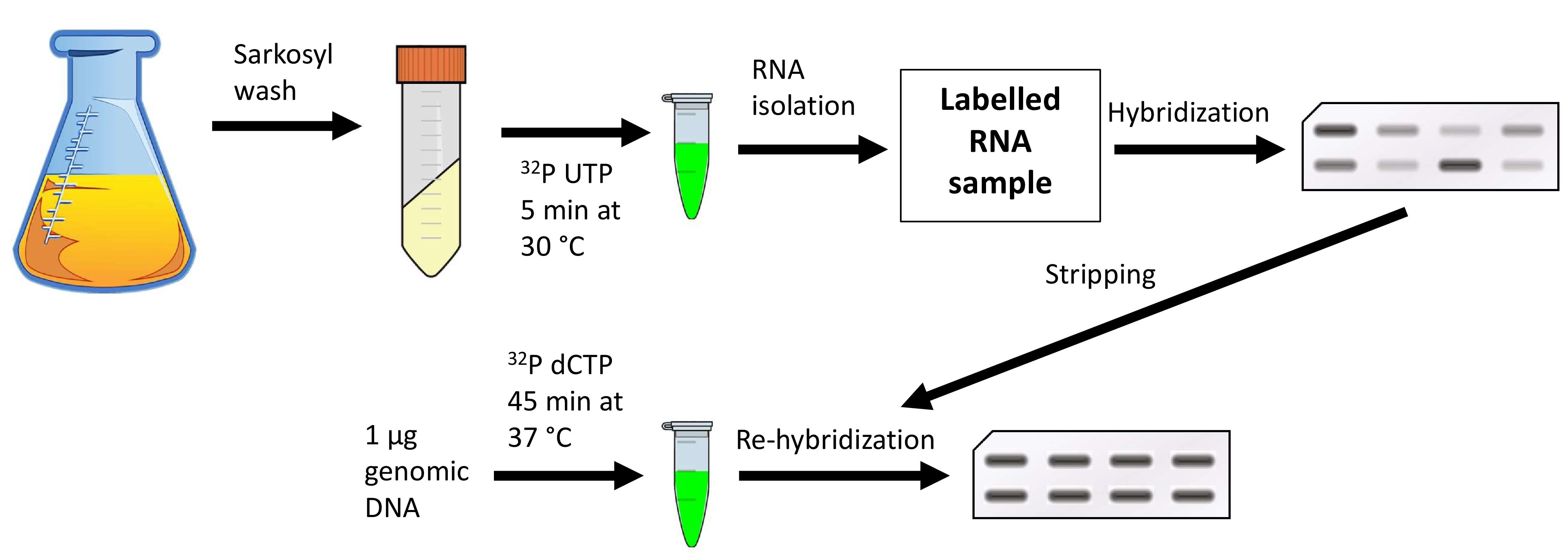
DNA transcription by RNA polymerases has always interested the scientific community as it is one of the most important processes involved in genome expression. This has led scientists to come up with different protocols allowing analysis of this process in specific locations across the genome by quantitating the amount of RNA polymerases transcribing that genomic site in a cell population. This can be achieved by either detecting the total number of polymerases in contact with that region (i.e., by chromatin immunoprecipitation (ChIP) with anti-RNA polymerase antibodies) or by measuring the number of polymerases that are effectively engaged in transcription in that position. This latter strategy is followed using transcription run-on (TRO), also known as nuclear run-on (NRO), which was first developed in mammalian cells over 40 years ago and has since been adapted to many other different organisms and high-throughput methods. Here, we detail the procedure for performing TRO in Saccharomyces cerevisiae for single genomic regions to study active transcription on a single gene scale. To do so, we wash the cells in the detergent sarkosyl, which prevents new initiations at the promoter level, and then perform an in situ reaction, leading to the radiolabeling of transcripts by RNA polymerases that were already engaged in transcription at the moment of harvesting. By subsequently quantitating the signal of these transcripts, we can determine the level of active transcription in a single gene. This presents a major advantage over other forms of transcription quantitation such as RNA polymerase ChIP, since in the latter, both active and inactive polymerases are measured. By combining both ChIP and TRO, the amount of inactive or paused polymerases on a particular gene can be estimated.
Graphic abstract:
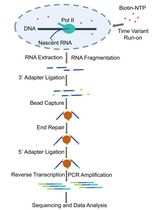
Transcriptional run-on scheme
Background
Transcription is an essential step in gene expression. It is a highly regulated process during which an RNA molecule is produced from a template DNA sequence by the action of RNA polymerases. Many scientists are interested in the complex process of transcription, which still remains to be fully understood and many others need to quantitate gene transcription in a specific region of the genome for practical purposes. Thus, several methods have been developed over the years to study eukaryotic transcription (reviewed in Pérez-Ortín et al., 2012).
Each developed method reveals different aspects of the transcription process. In this method article, we detail the protocol for transcription run-on (TRO), also known as nuclear run-on (NRO), which is particularly useful for detecting RNA polymerases that are engaged in active transcription at the moment of the experiment (Smale, 2009). The addition of sarkosyl detergent permeabilizes cells and prevents new initiations of transcription at the promoter level. Therefore, once new substrates (NTPs) are provided for the transcription reactions, only previously fully engaged RNA polymerases will act, incorporating a radiolabeled analog of UTP into the newly produced nascent RNA. This radiolabeling then allows for the detection of elongating RNA polymerases by hybridizing the nascent RNA onto nylon membranes that contain immobilized DNA fragments corresponding to the genomic sites of interest.
TRO was introduced over 40 years ago and has since experienced many upgrades for its use in different organisms and high-throughput sequencing (Greenberg and Ziff, 2004). The first attempt to take TRO genome-wide in yeast was the Genomic Run-On (GRO) method developed in 2004 (Garcia-Martinez et al., 2004), in which the labeled RNAs are hybridized to arrays. The protocol was then adapted to become high-resolution (Bio-GRO) and high-throughput (GRO-seq) (Core et al., 2008; Jordán-Pla et al., 2016 and 2019).
Here, we describe in detail the TRO method developed for Saccharomyces cerevisiae used in one of our recent publications (Corzo et al., 2019), which detects active RNA polymerases in single genes.
Materials and Reagents
Whatmann filter paper, 180 µm thickness (Macherey-Nagel, catalog number: 742213)
1.5 ml tubes (Eppendorf, catalog number: 22 36 411-1)
50 ml and 15 ml Falcon tubes (Eppendorf, catalog numbers: 0030122178 and 0030122151)
FastPrep 2 ml Lysing Matrix tubes (MP Biomedicals, catalog number: 115076200-CF)
ProbeQuantTM G-50 Micro Column (GE28-9034-08)
Saran wrap
Acetate film
Glass beads (Sigma, catalog number: G9268) washed in acid following the manufacturer’s protocol
HCl 37% (Sigma, catalog number: 320331)
Pfu DNA polymerase (Promega, catalog number: M7741)
QIAquick PCR Purification Kit (Qiagen, catalog number: 28104)
QIAquick Gel Extraction Kit (Qiagen, catalog number: 28115)
Yeast DNA Extraction Kit (Thermo Fisher, catalog number: 78870)
E. coli gDNA extraction (see He, 2011)
Hybond-N membrane (Amersham, catalog number: RPN303N)
NTP Set 100 mM (Invitrogen, catalog number: R0481)
Sarkosyl N-lauroylsarcosina (Sigma, catalog number: L5125)
DTT (Roche, catalog number: 11583786001)
α-32P UTP (Perkin-Elmer, catalog number: NEG507H001MC, 10 µCi/µl)
Cold MilliQ water
Acid phenol pH 4.3 (Sigma, catalog number: P4682)
Sodium acetate (Sigma, catalog number: 127-09-3)
100% ethanol (JT Baker, catalog number: 8025)
Chloroform (Sigma, catalog number: C2432)
NaOH pellets (Sigma, catalog number: 221465)
Random hexamers 50 µM (Invitrogen, catalog number: N8080127)
dNTP set (100 mM) (Invitrogen, catalog number: 10297018)
Klenow fragment (NEB, catalog number: M0212S)
α-32P dCTP (Perkin-Elmer, catalog number: NEG513H250UC, 10 µCi/µl)
NaCl (Labotaq, catalog number: SO0227005P)
Sodium citrate (Prolabo, catalog number: 27833.363)
MgCl2 (Prolabo, catalog number: 25108295)
EDTA (Sigma, catalog number: E5134-1KG)
SDS (Amresco, catalog number: 0227)
Sodium dihydrogen phosphate monohydrate (Sigma, catalog number: 10049-21-5)
Di-sodium hydrogen phosphate (Sigma, catalog number: 7558-79-4)
Potassium phosphate (Merck, catalog number: 1048731000)
Tris base (Sigma, catalog number: T1503-10KG)
Boric acid (Panreac, catalog number:131015.1211)
Yeast extract (Pronadisa, catalog number: 1702)
Glucose (Prolabo, catalog number: 24379.363)
Agarose low EEO (Sigma, catalog number: A0576-100G)
Red safe (Labotaq, catalog number: 8014)
DNA 1 Kb ladder (Invitrogen, catalog number: 10787-026)
DNA gel loading dye 6× (Thermo Scientific, catalog number: R0611)
Denaturalization buffer (see Recipes)
Neutralization buffer (see Recipes)
SCC 20× (see Recipes)
Transcription buffer 2.5× (see Recipes)
TES (see Recipes)
Hybridization solution (see Recipes)
Wash solution I (see Recipes)
Wash solution II (see Recipes)
Neutralization solution (stripping) (see Recipes)
Stripping solution (see Recipes)
10× TBE buffer (see Recipes)
Tris-HCl, pH 7 (see Recipes)
Phosphate buffer, pH 7 (see Recipes)
Equipment
Slot Blot blotting manifold (Hoefer Scientific, catalog number: PR648)
Thermocycler (Bio-Rad, catalog number: T100)
NanoDrop (Thermo Scientific, catalog number: 840274100)
Thermoblock heater (Labnet)
Vacuum pump
Spectrophotometer (Eppendorf, catalog number: EP6135000923)
Falcon centrifuge fixed rotor (Eppendorf, model: 5810R), max speed 3,220 × g
Microcentrifuge fixed rotor (Eppendorf, model: 5424), max speed 17,000 × g
UV crosslinker (Stratagene, model: UV Stratalinker 1800)
FastPrep-24TM 5G instrument (MP Biomedicals, catalog number: 116005500)
Hybridization oven (UVP, model: HB-1000 Hybridizer)
PhosphoImager screen and cassette (FUJIFILM BAS)
STORM-840 imaging system (GE Healthcare)
Vortex mixer (Jencons, model: VX100), use at max speed
Geiger counter (Thermo, model: Mini 900)
Microwave oven
Orbital shaker (Appleton Woods Stuart), slow shaking at about 50 rpm
Heat-resistant gloves
Electrophoresis chamber and power supply (Bio-Rad, catalog number: 1640300)
Gel casting tray and comb (Bio-Rad)
Software
GelQuant.NET software (http://biochemlabsolutions.com/GelQuantNET.html)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Begley, V., de Miguel-Jiménez, L. and Chávez, S. (2021). Transcriptional Run-on: Measuring Nascent Transcription at Specific Genomic Sites in Yeast. Bio-protocol 11(12): e4064. DOI: 10.21769/BioProtoc.4064.
分类
微生物学 > 微生物遗传学 > RNA > 转录
分子生物学 > RNA > 转录
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link