- EN - English
- CN - 中文
Cell-free Synthesis of Correctly Folded Proteins with Multiple Disulphide Bonds: Production of Fungal Hydrophobins
有多重二硫化物键的正确折叠蛋白质的无细胞合成:真菌疏水性化合物的生产
发布: 2021年05月20日第11卷第10期 DOI: 10.21769/BioProtoc.4019 浏览次数: 5061
评审: Alba BlesaRalph Thomas BoettcherMurugappan Sathappa
Abstract
Cell-free synthesis is a powerful technique that uses the transcriptional and translational machinery extracted from cells to create proteins without the constraints of living cells. Here, we report a cell-free protein production protocol using Escherichia coli lysate (Figure 1) to successfully express a class of proteins (known as hydrophobins) with multiple intramolecular disulphide bonds which are typically difficult to express in a soluble and folded state in the reducing environments found inside a cell. In some cases, the inclusion of a recombinant disulphide isomerase DsbC further enhances the expression levels of correctly folded hydrophobins. Using this protocol, we can achieve milligram levels of protein expression per ml of reaction. While our target proteins are the fungal hydrophobins, it is likely that this protocol with some minor variations can be used to express other proteins with multiple intramolecular disulphide bonds in a natively folded state.
Graphic abstract:

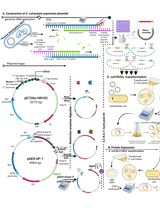
Figure 1. Workflow for cell-free protein expression and single-step purification using affinity chromatography. A. E. coli S30 lysate prepared as described in Apponyi et al. (2008) can be stored for up to several years at -80°C without any loss of activity in our experience. B. The S30 lysate, plasmid DNA that encodes for the protein of interest along with an affinity tag and components required for transcription and translation are added to the reaction mix. Following a single-step protein purification, the protein of interest can be isolated for further use.
Background
The bacterial cytosol microenvironment is home to thousands of diverse reactions and is tightly regulated in a reducing redox state. This poses challenges in recombinant bacterial protein overexpression as the reducing environment can prevent the correct formation of critical disulphide bonds in proteins that require them for structure and/or function (Kadokura and Beckwith, 2010). As such, these proteins are often expressed by E. coli into insoluble inclusion bodies (Berkmen, 2012) that renders them unusable. In favorable cases, proteins from inclusion bodies can be refolded in vitro after solubilizing in strong denaturants (Qin et al., 2015). However, this is typically a lengthy and reagent consuming process and the yield of the correctly refolded protein is variable.
One alternative to conventional recombinant bacterial overexpression is cell-free synthesis, where proteins can be expressed using the bacterial transcription and protein production machinery, but without the confines of a cell (Apponyi et al., 2008; Ozawa and Loh, 2014; Gao et al., 2019; Dondapati, 2020). Cell-free protocols are particularly useful in the expression of cysteine-rich proteins, as the redox environment can be controlled by simply changing the ratios of reduced and oxidized glutathione (GSH:GSSG) and varying the concentration of reducing agents such as dithiothreitol (DTT). After optimization, the yield of protein with correctly formed disulphide bonds suitable for downstream experiments (e.g., binding studies, structural and biophysical characterisation) can be greatly increased.
Beyond proteins with disulphide bonds, cell-free protein synthesis systems can hold additional advantages over traditional recombinant protein production such as the ability to readily incorporate unnatural amino acids (Gao et al., 2019; Dondapati et al., 2020). In addition, proteins that are too toxic for a host cell can be expressed in a cell-free environment as there is no requirement to maintain cell viability and this has allowed high throughput production and screening for membrane proteins, enzymes, and other therapeutics (Khambhati et al., 2019).
Our optimized cell-free expression protocol allows direct control of the redox conditions in the reaction mixture and demonstrates a distinct advantage over recombinant expression for a class of proteins known as the fungal hydrophobins (Siddiquee et al., 2020). This class of proteins is characterized by four disulphide bonds and the correct bonding pattern is required for hydrophobins to self-assemble into amphipathic layers at hydrophilic:hydrophobic interfaces (Sunde et al., 2008; Ball et al., 2020). These unique assemblies have suggested a range of biotechnological applications from drug delivery systems to coating implants and the engineering of surfaces to increase biocompatibility (Wosten and Scholtmeijer, 2015; Berger and Sallada, 2019). While many hydrophobins can be overexpressed in E. coli to a very high yield, the protein is almost exclusively found misfolded within inclusion bodies and cannot be used for downstream applications. Therefore, a long process of in vitro refolding and multi-step purification is typically required to separate the correctly folded protein from the partially folded and misfolded species (Kwan et al., 2006; Pille et al., 2015; Pham et al., 2016). To overcome this problem, we have developed a cell-free expression system for hydrophobin production based on the published S30 E. coli lysate (Apponyi et al., 2008).
The S30 lysate is one of the most used extracts in cell free synthesis of proteins and contains the cellular transcriptional and translational machinery. A Pubmed search with “S30 cell free” as search terms gives >150 articles. While the S30 lysate can be purchased commercially (e.g., from Promega), it can be produced readily in laboratories that perform recombinant protein expression and purification at a fraction of the cost of commercially available cell-free expression kits. This cell-free reaction involves incubating the lysate and other key cofactors with a gene of interest cloned downstream of a constitutive promoter in a dialysis set up. The dialysis set up allows continuous exchange of lower molecular reagents (e.g., amino acids, rNTPs, ATP) and offers higher protein yields per unit of components that are the more expensive or time-consuming to prepare (e.g., transcription and translation machinery in the S30 extract as well as plasmid DNA).
To aid purification, we have cloned the gene into the vector pET-MCSIII that has a hexa-histidine affinity tag but lacks the lac operator (Neylon et al., 2000). The protocol presented herein can be easily amended to enable the expression of other proteins with disulphide bonds. The apparatus used in this protocol can be easily prepared using disposable plastic tubes commonly used in most scientific laboratories and can be scaled up as required. The efficiency and low cost of this set up allow multiple proteins (e.g., a protein and its mutants) to be produced in parallel cell-free reactions using one preparation of dialysis buffer. Isotopically labeled proteins can also be easily produced by substituting in labeled amino acids in the reaction mixture. In some cases, this has allowed Nuclear Magnetic Resonance (NMR) and mass spectroscopy experiments to be carried out on the protein of interest within one day of setting up the reaction and without the need for further purification.
Materials and Reagents
Eppendorf tubes 1.5 ml (Thermo Fisher Scientific, catalog number: 69715)
Eppendorf tubes 0.5 ml (Eppendorf, catalog number: 0030121023)
Greiner Bio-One 50 ml tubes
Pipette tips
S30 Lysate prepared according to Apponyi et al. (2008)
rNTP (Sigma-Aldrich, catalog numbers: A6559, C8552, G3776, U1006)
HEPES (Sigma-Aldrich, catalog number: 15630106)
Folinic acid (Sigma-Aldrich, catalog number: 47612)
cAMP (Sigma-Aldrich, catalog number: A6885)
NH4OAc (Sigma-Aldrich, catalog number: A1542)
ATP (Sigma-Aldrich, catalog number: A1852)
Creatine phosphate (Sigma-Aldrich, catalog number: 27920)
Potassium glutamate (Sigma-Aldrich, catalog number: G1501)
Creatine kinase (Sigma-Aldrich, catalog number: C9983)
Mg(OAc)2 (Sigma-Aldrich, catalog number: M5661)
tRNA (Sigma-Aldrich, catalog number: R8759)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632)
L-Glutathione reduced (GSH) (Sigma-Aldrich, catalog number: G4251)
L-Glutathione oxidised (GSSG) (Sigma-Aldrich, catalog number: G4376)
20 amino acids (Sigma-Aldrich)
10,000 MWCO SnakeSkin® dialysis membrane (Thermo Fisher Scientific, catalog number: 88243). Please note to choose a membrane cutoff that is at least three times smaller than the size of the expected protein product
NuPAGETM 4-12% Bis-Tris Protein Gel (Invitrogen, catalog number: NP0323PK2) or any SDS-PAGE gel that is suitable for the detection of the protein of interest
MES buffer (Invitrogen, catalog number: NP0002) or other compatible buffers for the running the SDS-PAGE gel
NuPAGETM LDS Sample Buffer (4×) (Thermo Fisher Scientific, catalog number: NP0007)
PBS (Sigma-Aldrich, catalog number: P4417)
Urea (Sigma-Aldrich, catalog number: U5378)
Recombinant disulphide isomerase DsbC (Note 2)
Plasmid or PCR-amplified fragment encoding for the protein of interest (Note 3)
Equipment
Standard laboratory equipment commonly available in molecular biology and protein laboratories, such as pipettes, fridges, magnetic stirrers have not been included. Equipment that have similar specifications to those listed below can also be used.
Scalpel
NanoDropTM UV-Vis Spectrophotometer (Thermo Fisher Scientific, catalog number: ND-2000)
Bunsen burner
Incubator (Eppendorf, model: Innova 44; M1282-0002)
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) apparatus (ThermoFisher Scientific, catalog number: A25977)
Software
Microsoft Excel
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Siddiquee, R. and Kwan, A. H. (2021). Cell-free Synthesis of Correctly Folded Proteins with Multiple Disulphide Bonds: Production of Fungal Hydrophobins. Bio-protocol 11(10): e4019. DOI: 10.21769/BioProtoc.4019.
分类
生物化学 > 蛋白质 > 表达
生物化学 > 蛋白质 > 分离和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
提问指南
+ 问题描述
写下详细的问题描述,包括所有有助于他人回答您问题的信息(例如实验过程、条件和相关图像等)。
Share
Bluesky
X
Copy link