- EN - English
- CN - 中文
Proteomics Profiling of S-sulfurated Proteins in Acinetobacter baumannii
鲍曼不动杆菌硫酸化蛋白的蛋白质组学分析
发布: 2021年05月05日第11卷第9期 DOI: 10.21769/BioProtoc.4000 浏览次数: 5131
评审: Emilia KrypotouMilos FilipovicAnonymous reviewer(s)
Abstract
Hydrogen sulfide (H2S) is emerging as an important modulator in bacterial cytoprotection against the host immune response in infected animals, which may well be attributed to downstream highly oxidized sulfur species, termed reactive sulfur species (RSS), derived from H2S. One mechanism by which H2S/RSS may signal in the cell is through proteome S-sulfuration (persulfidation), which is the conversion of protein thiols (-SH) to protein persulfides (-SSH). While several analytical methods have been developed to profile sites of protein persulfidation, few have been applied to bacterial cells. The analytical workflow presented here was recently utilized to profile proteome persulfidation in the major human pathogen Acinetobacter baumannii treated with an exogenous sulfide source, Na2S. The data obtained using this protocol allow quantitation of the change in persulfidation status of each cysteine in the proteome normalized to the change in protein abundance, thus identifying sites of persulfidation that may constitute regulatory modifications. These can be validated using follow-up biochemical studies.
Keywords: Protein persulfidation (蛋白质过硫化/硫巯基化修饰)Background
To combat the increasing threat of drug-resistant and life-threatening pathogens, new antimicrobial strategies must be developed. One important approach to achieving this is to understand bacterial adaptation to the host immune response. Recent studies that build on prior work (Shatalin et al., 2011) suggest that bacterial H2S biogenesis may well be a clinically important adaptive response during infection (Mironov et al., 2017; Shukla et al., 2017; Luhachack et al., 2019; Toliver-Kinsky et al., 2019; Saini et al., 2020). The beneficial traits attributed to H2S are likely due to highly oxidized sulfur species termed reactive sulfur species (RSS), e.g., organic persulfides (RSSH), which have been shown to be potent antioxidants in mammalian cells (Ida et al., 2014). Recently, RSS has been quantitated in a number of different bacterial pathogens, suggesting that the antioxidant and cytoprotective findings in mammalian cells may well extend to bacterial cells (Peng et al., 2017a and 2017b; Shen et al., 2018; Walsh et al., 2020).
The mechanism by which H2S/RSS signals in the cell is not completely understood but almost certainly involves the protein post-translational modification (PTM) termed S-sulfuration (persulfidation) (Filipovic et al., 2018; Walsh and Giedroc, 2020). This PTM may provide protection against over-oxidation of protein thiols or perform a regulatory role by altering the chemistry of active-site thiols, thiols in regulatory domains, or those found in transcriptional regulators. Investigating the regulatory nature of protein persulfidation is complicated by the fact that protein thiols can form a number of other redox modifications, e.g., S-sulfenylation (RSOH) or S-nitrosation (RSNO), rendering it difficult to distinguish those proteins for which persulfidation is regulatory versus a consequence of a highly reactive or surface-exposed thiol. Most published methods for the detection of protein persulfidation incorporate: i) streptavidin-based alkylation probes to enrich for thiols and persulfide-harboring peptides followed by selective reduction of the mixed disulfide originating from alkylated persulfides (Gao et al., 2015; Peng et al., 2017b); and/or ii) a tag-switch approach that further exploits the unique reactivity of this mixed disulfide toward a specific nucleophile (Zhang et al., 2014; Park et al., 2015; Zivanovic et al., 2019). Recently, low-pH quantitative thiol reactivity profiling (QTRP) was introduced to directly detect protein persulfides and thiols in the same sample, an important step toward defining the ratio of persulfide to thiol at all cysteines (Fu et al., 2020).
The proteomics workflow presented here was recently applied to the major human pathogen Acinetobacter baumannii, in which several hundred proteins were identified as sites of persulfidation in both untreated and Na2S-treated cells (Walsh et al., 2020). This method relies on the alkylation of both thiol and persulfide residues using a biotinylated iodoacetamide probe. The proteins are then digested into peptides and enriched using streptavidin bead capture, after which peptides containing a mixed disulfide that is derived only from persulfidated residues are selectively reduced, alkylated, and identified by LC-MS/MS (Figure 1A). Although false positives can result from the reaction of these electrophilic probes with sulfenylated cysteines (Reisz et al., 2013), lower concentrations of reagent and shorter alkylation times can minimize this. In addition, it is important to point out that H2S reacts directly with a sulfenylated cysteine to form a persulfide (Cuevasanta et al., 2015 and 2019); therefore, the extent to which sulfenylated cysteines would be present in a proteome following incubation of bacterial cell cultures with excess H2S is unknown, but may well be relatively low.
In a parallel experiment, protein abundance is determined for the entire bacterial lysate without enrichment using a widely used label-free quantitation method (Zhu et al., 2010). The two data sets are then used to calculate or simply compare the change in persulfidation status normalized to the change in protein abundance (Figure 1B), thus pinpointing cysteine thiols for which persulfidation may well be regulatory in nature. This workflow leads to the identification of over 40 proteins in A. baumannii characterized by a ≥2-fold change in the normalized persulfidation status, a significant number of which are transcriptional regulators, including two implicated in biofilm formation (Walsh et al., 2020).
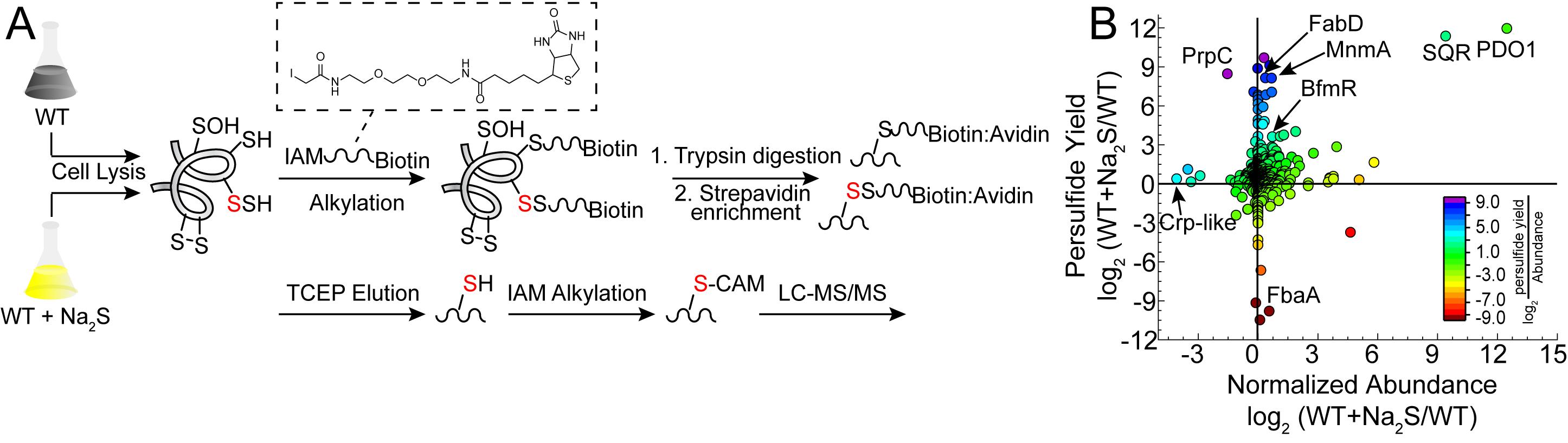
Figure 1. Proteome persulfidation profiling. A. General workflow for the enrichment and identification of proteome persulfides (Peng et al., 2017b). Both untreated (WT) and treated (WT+Na2S) samples are processed using the same workflow and analyzed independently of one another. B. Resulting data are plotted as a change in persulfidation status for a cysteine-containing peptide in a protein versus a change in the normalized abundance of that protein in WT vs. Na2S-treated cells (Walsh et al., 2020). Abbreviations: BfmR, biofilm response regulator; Crp-like, cAMP-activated regulator; FabD, ACP-malonyl transferase; FbaA, fructose-bisphosphate aldolase; MnmA, tRNA 2-thiouridine synthase; PDO1, persulfide dioxygenase-1; PrpC, 2-methylcitrate synthase; SQR, sulfide-quinone oxidoreductase.
Materials and Reagents
Note: Catalog numbers for specific suppliers that we use are given for many of the reagents listed below; an equivalent quality reagent from an alternative supplier can be substituted, with comparable results. Standard materials and reagents for cell culture are required and are not explicitly listed below. Reagents are listed in the approximate order of use in the protocol.
Culture tubes (VWR, catalog number: 60818-667)
50 ml centrifuge tubes (VWR, catalog number: 89039-656)
1.5 ml centrifuge tubes (VWR, catalog number: 20170-038)
Lysing Matrix B, 2 ml tubes (MP Biomedical, catalog number: 116911050)
2 ml centrifuge tubes (VWR, catalog number: 20170-170)
C18 Sep-Pak (Waters, catalog number: WAT051910)
Omix C18 tips (Agilent, catalog number: A57003100)
pH indicator paper
Acinetobacter baumannii (A. baumannii) ATCC 17978 strain
Luria-Bertani (LB) broth (Fisher, catalog number: BP1426)
Na2S (Sigma, catalog number: 407410)
Na2HPO4 (Fisher, catalog number: S374-500)
KH2PO4 (Fisher, catalog number: P285-500)
KCl (Sigma, catalog number: 7300-500GM)
NaCl (Macron, catalog number: 7581-06)
2-Morpholinoethanesulfonic acid, free acid (MES) (GoldBio, catalog number: M-095-1)
Liquid nitrogen
Bradford reagent (Thermo, catalog number: 23238)
Bovine serum albumin (BSA) (Sigma, catalog number: A4503)
3 kDa cut-off concentrators (Millipore Sigma, catalog number: UFC500396)
Urea (Fisher, catalog number: U15)
Ammonium bicarbonate (ABC) (J.T. Baker, catalog number: 3003-01)
EZ-LinkTM Iodoacetyl-PEG2-Biotin (Biotinylated iodoacetamide, BIAM) (Thermo, catalog number: 21334)
Sequencing grade trypsin (Promega, catalog number: V5117)
Avidin acrylamide resin (Thermo, catalog number: 53151)
Tris(2-carboxyethyl)phosphine (TCEP) (ChemImpex, catalog number: 23004)
Iodoacetamide (IAM) (Sigma, catalog number: I1149)
Trichloroacetic acid (TCA) (Sigma, catalog number: 91228)
Diluted solutions of organic acid (0.1-10%) commonly used in mass spectrometry, e.g., trifluoroacetic acid or formic acid
LC-MS grade acetonitrile (OmniSolv, catalog number: AX0156)
LC-MS grade water (OmniSolv, catalog number: WX0001-1)
Na2S stock (see Recipes)
Phosphate-buffered saline (PBS) (see Recipes)
Lysis buffer (see Recipes)
Sep-Pak Aqueous Buffer (see Recipes)
Sep-Pak Organic buffer (see Recipes)
Equipment
Note: Typical biochemistry lab equipment is required; therefore, it is not explicitly listed below. Catalog numbers are given for most of the equipment as examples; instruments from alternative manufacturers may be substituted, provided equivalent functionality. Equipment is listed in the approximate order of use in the protocol.
Shaker
Spectrophotometer and cuvettes
Various centrifuges with the appropriate rotors capable of centrifuging a variety of volumes ranging from 1.5 ml to 50 ml or greater (depending on the cell culture volume)
Water bath
Bead Ruptor 12 Homogenizer (Omni, model: 19-050A)
Anaerobic chamber (Vacuum Atmospheres Company, model: HE-453-2-245)
Rotor-Barnstead Thermolyne Labquake Shaker (Barnstead International, model: 400110)
Vacuum manifold (Thermo Fisher Scientific, model: 60104-232)
ISS 110 SpeedVac System (Thermo Fisher Scientific, model: ISS 110)
Trapping column: Symmetry C18 trap column (180 μm × 20 mm, 5 μm)
Analytical column: nanoACQUITY HPLC HHS T3 analytical column (75 μm × 150 mm, 1.8 μm)
Orbitrap FusionTM LumosTM TribridTM Mass Spectrometer (Thermo Fisher Scientific) interfaced with a nano-flow liquid chromatography (LC) system (e.g., Thermo Scientific EASY-nLC 1200)
Software
UniProt
Proteome Discoverer 2.1 (Thermo Scientific)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Walsh, B. J. C. and Giedroc, D. P. (2021). Proteomics Profiling of S-sulfurated Proteins in Acinetobacter baumannii. Bio-protocol 11(9): e4000. DOI: 10.21769/BioProtoc.4000.
分类
生物化学 > 蛋白质 > 翻译后修饰
微生物学 > 微生物蛋白质组学 > 全生物体
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













