- EN - English
- CN - 中文
AIMTOR, a BRET Biosensor for Live Recording of mTOR Activity in Cell Populations and Single Cells
一种用于实时记录细胞群和单个细胞中mTOR活性的BRET生物传感器AIMTOR
(*contributed equally to this work) 发布: 2021年04月20日第11卷第8期 DOI: 10.21769/BioProtoc.3989 浏览次数: 4450
评审: Joachim GoedhartSalma MerchantAnonymous reviewer(s)
Abstract
Mammalian target of rapamycin (mTOR) controls many crucial cellular functions, including protein synthesis, cell size, energy metabolism, lysosome and mitochondria biogenesis, and autophagy. Consequently, deregulation of mTOR signaling plays a role in numerous pathological conditions such as cancer, metabolic disorders and neurological diseases. Developing new tools to monitor mTOR spatiotemporal activation is crucial to better understand its roles in physiological and pathological conditions. However, the most widely used method to report mTOR activity relies on the quantification of specific mTOR-phosphorylated substrates by western blot. This approach requires cellular lysate preparation, which restricts the quantification to a single time point. Here, we present a simple protocol to study mTOR activity in living cells in real time using AIMTOR, an intramolecular BRET-based (bioluminescence resonance energy transfer) biosensor that we recently designed (Bouquier et al., 2020). We describe transfection of AIMTOR in the C2C12 cell line and procedures to monitor BRET in a cell population using a plate reader and in single cells by microscopy. Importantly, this protocol is transposable to any cell line and primary cells. In addition, several subcellular compartment-specific versions of AIMTOR have been developed, enabling compartmentalized assessment of mTOR activity. This protocol describes how to use the sensitive AIMTOR biosensor to investigate mTOR signaling dynamics in living cells.
Graphic abstract:
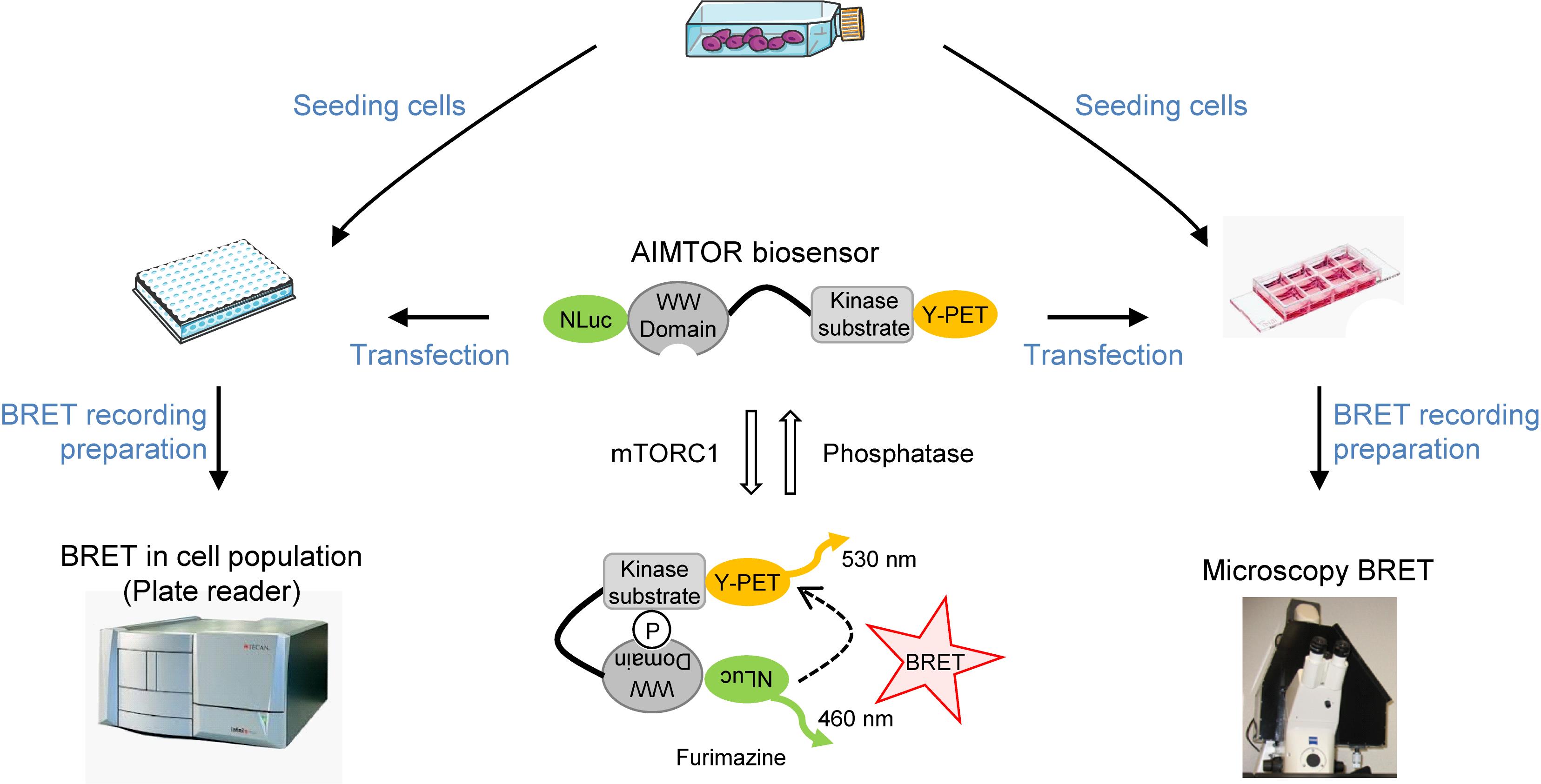
AIMTOR protocol overview from seeding cells to live BRET recording
Background
The mTOR signaling pathway is conserved within the eukaryotic domain and controls major metabolic and anabolic pathways involved in cellular homeostasis, including proteostasis, lipogenesis, and glucose and mitochondrial metabolism. The mTOR kinase occupies the central stage of this signaling pathway and its activation/inhibition depends on a myriad of environmental cues, including nutrient availability, growth factors, energy status and various stresses. mTOR signaling is regulated by complex and sometimes redundant mechanisms depending on cellular type and context. As expected given its central role in cellular homeostasis, many if not all human physiopathological conditions involve mTOR signaling to some degree or will at least interfere with its activation/regulation at some stage of their progression. The mTOR kinase belongs to several multiprotein complexes, including mTORC1 and mTORC2, which exhibit distinct cellular functions and substrate specificities. The mTORC1 complex primarily controls cell growth and anabolism pathways, whereas the mTORC2 complex is involved in regulation of cellular metabolism (Saxton and Sabatini, 2017). Efforts are underway to better investigate mTOR signaling and activation modes in several cell types. To this end, specific tools are required to accurately monitor mTOR kinase activity in living cells in real time.
However, current methods mostly rely on endpoint approaches requiring cell lysis, precluding dynamic studies of live cells and kinetic investigations. For example, western blot analysis requires quantification of the abundance of several specific endogenous substrates phosphorylated by mTOR in cell or tissue protein lysates. To circumvent this drawback, genetically encoded biosensors have been designed that allow kinase activity to be monitored in living cells. Indeed, fluorescence resonance energy transfer (FRET) biosensors have been developed to allow imaging of the activity of several important signaling kinases in living cells, including ERK, S6K, AKT, and mTOR. These intramolecular FRET biosensors monitor conformational change-induced shifts in the proximity between donor and acceptor fluorophores upon phosphorylation of a specific kinase substrate. However, drawbacks related to the use of excitation light, such as cell auto-fluorescence, spectral overlaps between donor and acceptor excitation and fluorophore bleaching, complicate the analysis and interpretation of FRET data. Because it does not require light excitation, bioluminescence resonance energy transfer (BRET) circumvents the major technical limitations of FRET. The donor for BRET is a luciferase entity that emits light only after chemical activation. BRET has been shown to be an efficient approach not only for protein/protein interaction studies, but also for the design of biosensors to monitor enzymatic activity.
Here, we describe a protocol in which the genetically encoded BRET biosensor AIMTOR (Bouquier et al., 2020) is used to monitor mTOR cell signaling by BRET imaging in individual cells under a microscope or in cell populations with a plate reader. This detailed protocol was recently validated to investigate mTORC1 signaling and dysfunction in muscle cells and primary neurons (Bouquier et al., 2020). However, AIMTOR can be used in virtually all types of cultured cells, cell lines or other primary cells. AIMTOR enables delineation of mTOR signaling, identification of activators and inhibitory conditions, and screening of drugs/compounds that interfere with this important cellular hub. In addition, several versions of AIMTOR targeted to various subcellular compartments have been designed to better characterize the existence of discrete compartmentalized pools of mTOR activity. Future versions of AIMTOR utilizing new vectors containing tissue-specific promoters or other subcellular tags will expand the range of methods available for monitoring mTOR signaling directly in live cells and organs.
Materials and Reagents
100 mm TC-treated Culture Dish (Corning, catalog number: 430167 ) and TTP 6-well plates (Sigma, catalog number: Z707759 )
8-well IbiTreat µ-Slide (ibidi, catalog number: 80826) or 8-well Polylysine-coated Slides (ibidi, catalog number: 80824)
96-well clear bottom white microplate (Greiner, catalog number: 655098 )
1.5 ml and 15 ml tubes
C2C12 mouse myoblast cell line (ATCC, catalog number: CRL-1772 )
AIMTOR plasmid DNA T757 (Addgene ID 140828) and nonphosphorylatable A757 mutant (Addgene ID 140829), store at -20 °C
Dulbecco’s Modified Eagle’s Medium (Sigma, catalog number: D5796-500ml ), store at 4 °C
Dulbecco’s Modified Eagle’s Medium (Gibco, without phenol red, glutamine and glucose A1443001-500 ml), store at 4 °C
Glucose 20% (1.11 M), filtered and sterilized; store at 4 °C. Glucose 20% is used to complement phenol red-free and glucose-free DMEM with 25 mM glucose.
Fetal bovine serum (Dominique Dutscher, catalog number: S1810-500), store at -20 °C
Glutamine 200 mM (Sigma, catalog number: G7513-100ml ), store at -20 °C
Na pyruvate 100 mM (Sigma, catalog number: S8636-100ml), store at 4 °C
Antibiotics penicillin and streptomycin, 10,000 U/ml (Sigma, catalog number: P4333 ), store at -20 °C
HEPES buffer, 1 M, filtered and sterilized (Sigma, catalog number: H3662 ), store at 4 °C
PBS (Sigma, catalog number: D8537-500 ml), store at room temperature
Trypsin/EDTA (Sigma, catalog number: T4049-100ml ), store at 4 °C
Furimazine (Promega, Nano-Glo luciferase assay, catalog number: N1120 ), store at -20 °C
JETPEI (Polyplus Transfection Reagent, catalog number: 101-10N ), store at 4 °C
Helix-IN DNA transfection reagent (OZBiosciences, catalog number: HX10100, store at -20 °C) or any agent to efficiently transfect your cells of interest without affecting cell phenotype and viability
DMEM regular growth medium (contains phenol red) (see Recipes)
DMEM growth medium and reading medium (see Recipes)
Equipment
MicroPipettes
Multicanal pipette (10-200 µl) and/or repipetor with 500 µl-10 µl tip (Eppendorff)
Water-jacketed CO2 incubator
Class II biological safety cabinet
Thoma counting chamber or other cell counting device
Centrifuge (SIGMA, model: 3-16KL ) with a rotor (SIGMA, catalog number: 11222) suitable for 6-well plates (for JETPEI transfection)
For BRET population measurement
SynergyTM 2 Biotek Microplate reader (or an equivalent microplate reader) allowing sensitive measurement of bioluminescence filtered emission light around 460 nm (emission peak of nanoluciferase induced by furimazine) and 530 nm (emission peak of YPET)
Emission filters: 485/20 nm (Biotek, catalog number: 7082221) and 530/25 nm (Biotek, catalog number: 7082223)
Gen5TM Reader Control and Data Analysis Software to program the plate reader and generate raw data files (in Excel, Microsoft OfficeTM)
For BRET imaging
Homemade dark box with sliding trap (Figure 1A)
Computer with Metamorph software (Figure 1B)
Bioluminescence-dedicated microscope device customized from an inverted fluorescence microscope (Carl Zeiss, Axiovert 200M) (Coulon et al., 2008; Perroy, 2010) (Figure 1C). Please note that due to the strong brightness of NanoLuc, it is now possible to use a classic inverted microscope (Kim and Grailhe, 2016)
Evolve camera equipped with an EMCCD detector, back-illuminated, with On-chip Multiplication Gain (Photometrics) (Figure 1D)
Multiposition stage control (Marzhauser Wetzlar, Tango 2 Desktop, catalog number: 00-76-150-1802) (Figure 1E)
Illumination source: Lumencor (Gataca, Spectra X light engine) (Figure 1F)
Plan-Neofluar 40×/1.3 oil objective M27 (Carl Zeiss)
Emission filters: 450/70 nm (Semrock, catalog number: FF01-450/70-25) and HQ535/50nm (Chroma, catalog number: 63944)
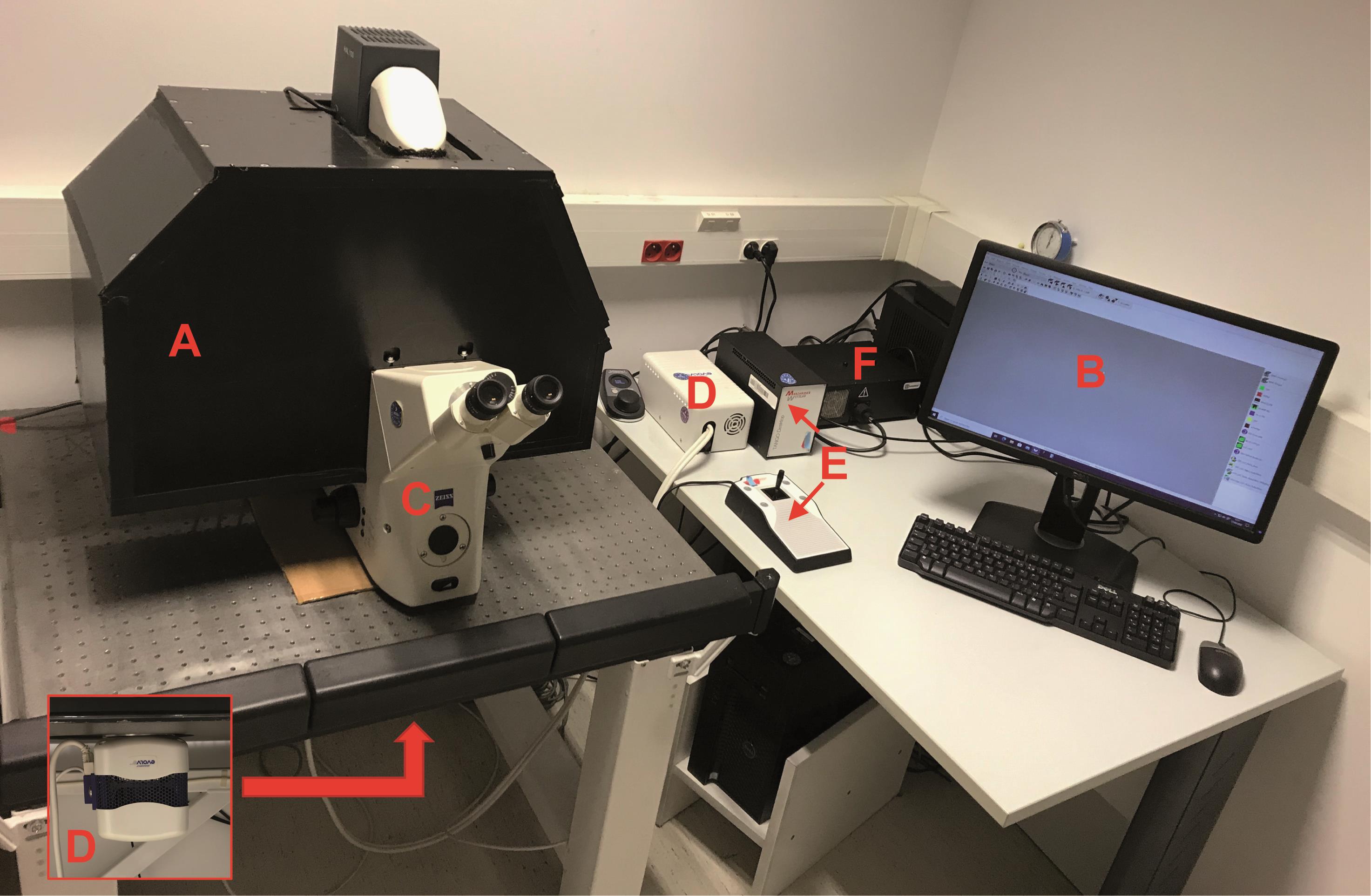
Figure 1. Microscopy BRET imaging setup. A. Dark box. B. Computer. C. Microscope. D. Camera. E. Multiposition stage control. F. Light source.
Software
Gen5TM Reader Control and Data Analysis Software for Biotek microplate reader
Metamorph image acquisition software (Molecular Devices)
FIJI (Schindelin et al., 2012)
BRET analyzer FIJI plugin (Chastagnier et al., 2018) https://github.com/ychastagnier/BRET-Analyzer
Graphpad Prism 7 software (GraphPad Software Inc.)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Bouquier, N., Moutin, E., Perroy, J. and Ollendorff, V. (2021). AIMTOR, a BRET Biosensor for Live Recording of mTOR Activity in Cell Populations and Single Cells. Bio-protocol 11(8): e3989. DOI: 10.21769/BioProtoc.3989.
分类
细胞生物学 > 细胞信号传导 > 磷酸化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












