- EN - English
- CN - 中文
Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Live-cell Vesicle Tracking in the Secretory Pathway Using HeLa Cells
用RUSH蛋白分选法追踪HeLa细胞分泌通路中的活细胞囊泡
(*contributed equally to this work) 发布: 2021年03月20日第11卷第6期 DOI: 10.21769/BioProtoc.3958 浏览次数: 5239
评审: Begoña DiazAnonymous reviewer(s)
Abstract
More than 30% of the total amount of proteins synthesized in mammalian cells follow the secretory pathway in order to mature and be properly sorted to their final destinations. Among several methodologies that describe live-cell monitoring of vesicles, the Retention Using Selective Hooks (RUSH) system is a powerful one that allows to visualize cargo trafficking under physiological conditions. The present protocol describes a method to use the RUSH system in live-cell microscopy and a subsequent quantitative analysis of cargo vesicles to dissect protein trafficking. In brief, HeLa cells are transiently transfected with an MMP2-RUSH construct and vesicle trafficking is evaluated by wide-field microscopy, recording videos in 1-min time frames for 45 min. We also present a quantitative approach that can be used to identify kinetics of uncharacterized protein cargo, as well as to evaluate with more detail processes such as ER-to-Golgi vesicle trafficking.
Graphic abstract:
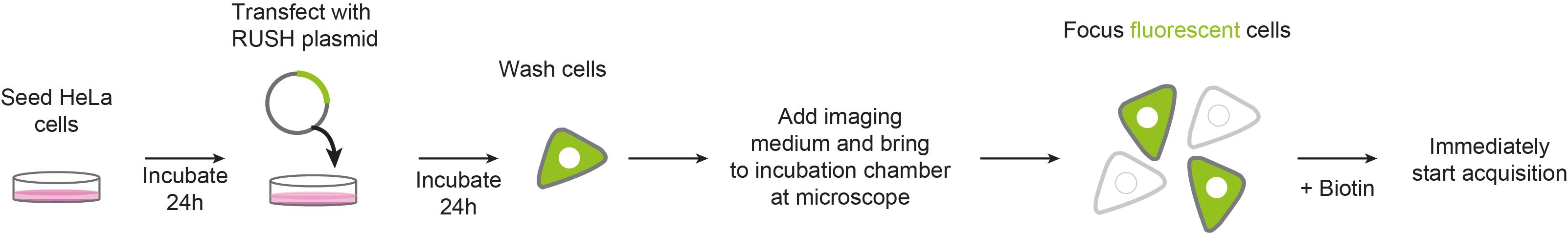
Live-cell RUSH: a tool to monitor real-time protein trafficking in the secretory pathway
Background
More than 30% of the total amount of proteins synthesized in mammalian cells follow the secretory pathway (Pfeffer, 2010; Boncompain and Weigel, 2018). Through this pathway, proteins mature by trafficking from the ER and along the different Golgi stacks until they reach the trans-Golgi network (TGN), where they are finally sorted and packed into vesicles that will be delivered to other organelles within the cell, or to the extracellular milieu (Glick and Luini, 2011; Pantazopoulou and Glick, 2019).
In the recent years, several reports in the literature document advance in the live-cell monitoring of vesicles that follow the secretory pathway (Stephens and Perez, 2013). One of the most helpful tools is the Retention Using Selective Hooks (RUSH) system, a methodology that enables the synchronized monitoring of fluorescent vesicles in a living cell upon biotin addition (Boncompain et al., 2012). In this system, cells are either transiently transfected or stably express a two-part protein complex consisting of a protein of interest (POI) tagged with a fluorophore and bound to a streptavidin binding peptide (SBP), and streptavidin bound to a retention signal (known as the “hook”, e.g., a KDEL sequence for retention in the ER; Figure 1). Given the higher affinity of streptavidin to biotin, once the latter is added to the cell culture media, the SBP-POI complex is released to the next compartment and the trafficking can be followed either live by wide-field microscopy or monitored at different fixed time points by confocal microscopy (Boncompain et al., 2012).
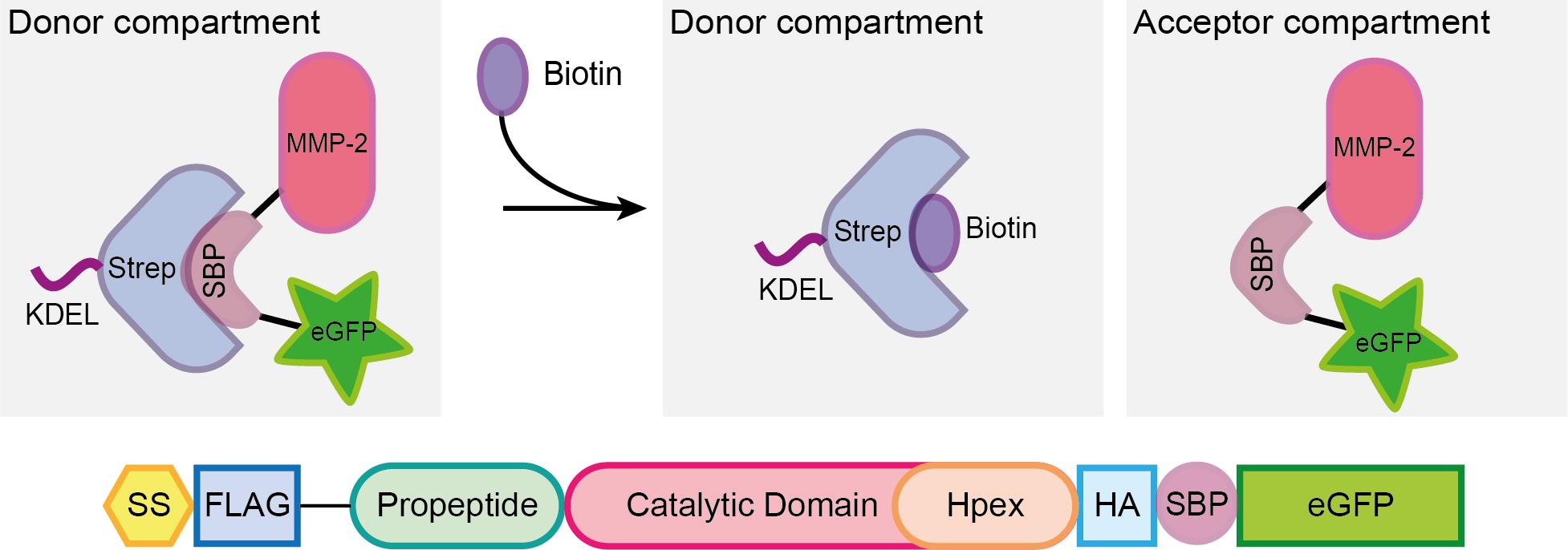
Figure 1. Scheme representing the RUSH system. A protein complex containing the protein of interest (here matrix metalloprotease 2, MMP2), a streptavidin binding peptide (SBP) and a fluorescent protein (eGFP) is bound via SBP to streptavidin (Strep), which is linked to a KDEL sequence for the retention of the complex in the donor compartment (in this example, the ER). Once biotin is added to the media, it binds to streptavidin, enabling the MMP2-SBP-eGFP protein complex to travel to the acceptor compartment (the Golgi apparatus in the secretory pathway). Figure taken from Pacheco-Fernandez et al. (2020) and adapted from Boncompain et al. (2012).
One of the main advantages of using the RUSH system is that it enables to carefully dissect the trafficking kinetics of soluble and membrane proteins that follow the secretory pathway under physiological conditions (Boncompain et al., 2012). Some other techniques developed in recent years have also enabled the characterization of vesicle trafficking through the secretory pathway. Among others, super-resolution confocal live imaging (Kurokawa et al., 2019); synchronization of cargo-retention and release by polymerization/depolymerization (Cancino et al., 2013) and a light-triggered protein secretion system (Chen et al., 2013). However, such techniques require either a more complex set up, the use of more sensitive equipment, or the aggregation of the protein of interest previous to its synchronized trafficking, which may lead to cargo trafficking through different pathways than those followed by the unaggregated protein (Boncompain and Perez, 2013).
Based on the original RUSH methodology developed by Boncompain et al. (2012), here we present a protocol that provides in addition a quantitative analysis, which aims to better dissect intra-Golgi trafficking by counting the number of cargo vesicles in 1-minute time video frames during a defined period of time, in order to assess post-Golgi cargo sorting kinetics (Deng et al., 2018). Furthermore, the ImageJ macro developed for such analysis is also optimized for dissecting the ER-to-Golgi cargo trafficking kinetics using Golgi compaction (ratio between Golgi:ER fluorescent signal per minute) as a measure (Pacheco-Fernandez et al., 2020).
Materials and Reagents
35 mm µ-dish, High Glass Bottom (ibidi, catalog number: 81158 )
Polyethylenimine (PEI), linear, MW 25000, transfection grade (PEI 25KTM; Polysciences, catalog number: 23966-1 )
60-70% confluent HeLa cells
For this analysis, cells were obtained from Cell Lines Service. We used these cells because they are easy to transfect, have a good size for vesicle visualization and are easy to handle, however, other cell lines can also be used (see Note 1).
1× Dulbecco’s Phosphate Buffered Saline (DPBS), no calcium, no magnesium (Life Technologies, Gibco, catalog number: 14190144 ). Storage temperature: 4 °C
pIRESneo3-Str-KDEL-MMP2-SBP-EGFP vector. Storage temperature: -20 °C
Important note: This plasmid was generated by replacing the ST (ST6GAL1) sequence by the MMP2 one (our protein of interest) in the Addgene plasmid number 65264 (Str-KDEL_ST-SBP-EGFP).
Opti-MEM® reduced serum media (Life Technologies, Gibco, catalog number: 31985070 ). Storage temperature: 4 °C
500 mM d-biotin (Merck, SUPELCO, catalog number: 47868 ). Storage temperature: 4 °C
Dulbecco’s Modified Eagles Medium, high glucose, GlutaMAXTM, sodium pyruvate (DMEM, Life Technologies, Gibco, catalog number: 10569010 ). Storage temperature: 4 °C
Heat inactivated Fetal Bovine Serum (FBS; Life Technologies, Gibco, catalog number: 16000044 ). Storage temperature: -20 °C
Penicillin/Streptomycin (P/S; Life Technologies, Gibco, catalog number: 15140122 ). Storage temperature: -20 °C
DMEM, high glucose, HEPES, no phenol red (Life Technologies, Gibco, catalog number: 21063029 ). Storage temperature: 4 °C
PEI solution at 1 mg/ml (see Recipes)
DMEM complete medium (see Recipes)
Equipment
Magnetic stirrer
Laminar flow hood
Cell incubator set at 37 °C, 5% CO2
Water bath set at 37 °C
Microscope:
DeltaVision Elite System based on:
Olympus IX-71 inverted microscope (Olympus Corporation)
Olympus 60×/1.42 PLAPON oil objective (Olympus Corporation)
PCO pco.edge sCMOS 5.5 microscope camera
7-colour InsightSSI module laser
DAPI-FITC-TRITC-Cy5 and/or CFP-YFP-mCherry filters
Live-cell imaging incubation chamber set up at 37 °C. Optional: 5% CO2 and humidity.
Software
SoftWoRx 5.5 software (GE Healthcare, https://cdn.cytivalifesciences.com/dmm3bwsv3/AssetStream.aspx?mediaformatid=10061&destinationid=10016&assetid=17238)
Fiji (Schindelin et al., 2012, https://fiji.sc)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Pakdel, M., Pacheco-Fernandez, N. and von Blume, J. (2021). Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Live-cell Vesicle Tracking in the Secretory Pathway Using HeLa Cells. Bio-protocol 11(6): e3958. DOI: 10.21769/BioProtoc.3958.
- Pacheco-Fernandez, N., Pakdel, M., Blank, B., Sanchez-Gonzalez, I., Weber, K., Tran, M. L., Hecht, T. K., Gautsch, R., Beck, G., Perez, F., Hausser, A., Linder, S. and von Blume, J. (2020). Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs. J Cell Biol 219(8).
分类
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












