- EN - English
- CN - 中文
Trypanosomatid, fluorescence-based in vitro U-insertion/U-deletion RNA-editing (FIDE)
基于荧光的锥虫体体外尿嘧啶插入或删除RNA编辑(FIDE)
发布: 2021年03月05日第11卷第5期 DOI: 10.21769/BioProtoc.3935 浏览次数: 4640
评审: Manuel Miras MarinKarolina SubrtovaMarcelo S. da Silva
Abstract
Gene expression within the mitochondria of African trypanosomes and other protozoan organisms relies on a nucleotide-specific RNA-editing reaction. In the process exclusively uridine (U)-nucleotides are site-specifically inserted into and deleted from sequence-deficient primary transcripts to convert them into translatable mRNAs. The reaction is catalyzed by a 0.8 MDa multiprotein complex termed the editosome. Here we describe an improved in vitro test to quantitatively explore the catalytic activity of the editosome. The assay uses synthetic, fluorophore-derivatized oligoribonucleotides as editing substrates, which enable the automated electrophoretic separation of the reaction products by capillary electrophoresis (CE) coupled to laser-induced fluorescence (LIF) detection systems. The assay is robust, it requires only nanogram amounts of materials and by using multicapillary CE/LIF-instruments it can be executed in a highly parallel layout. Further improvements include the usage of phosphorothioate-modified and thus RNase-resistant substrate RNAs as well as multiplex-type fluorophore labeling strategies to monitor the U-insertion and U-deletion reaction simultaneously. The assay is useful for investigating the mechanism and enzymology of the editosome. However, it can also be executed in high-throughput to screen for RNA editing-specific inhibitors.
Graphic abstract:
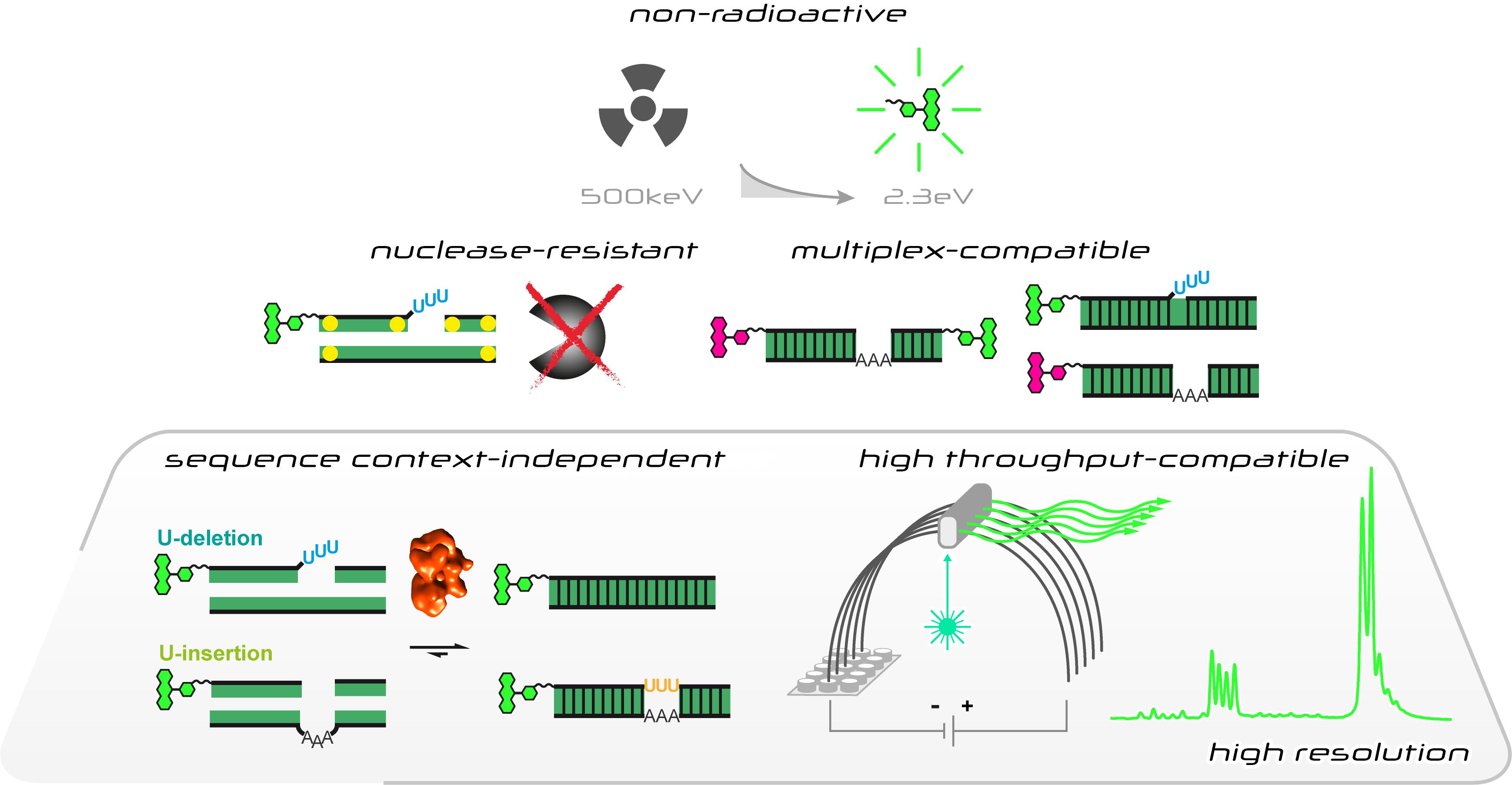
Characteristics of the fluorescence-based in vitro U-insertion/U-deletion RNA-editing (FIDE) assay
Background
The RNA editing reaction in kinetoplastid protozoa such as African trypanosomes and Leishmania represents one the most striking posttranscriptional modifications of messenger (m)RNAs (reviewed in Göringer, 2012a). The reaction takes place within the single mitochondria of the organisms, which is similar to other eukaryotes and holds the genetic information for components of the mitochondrial translation apparatus and subunits of the electron transport and chemiosmosis systems. However, unlike in other organisms, more than 50% of these genes are “incompletely” encoded: substantial sequence information is missing – in some cases more than 50% – in addition to the absence of start and stop codons and the presence of frame-shifts. As a consequence, precursor (pre)-mRNAs from these genes must be corrected, i.e., "edited" to generate functional mRNAs. Biochemically, the editing reaction is characterized by the insertion and/or deletion of U-nucleotides. In African trypanosomes, more than 3000 U-residues are inserted and about 300 U’s deleted at hundreds of editing sites in 12 different pre-mRNAs. The process relies on small, non-coding RNAs, known as guide (g)RNAs (Blum et al., 1990; Hermann et al., 1995). Guide RNAs are trans-acting genetic elements. They initiate the reaction cycle by antiparallel base pairing to the pre-edited mRNAs, which results in the formation of a gRNA/pre-mRNA hybrid. The hybrid-RNA has a 3-helix junction-topology including a so-called “anchor” – helix that borders the sequence to be edited. Unpaired gRNA nucleotides next to the anchor-helix specify U-insertion events, non-base paired uridylates in the pre-mRNA become deleted. The mitochondrial machinery that executes the processing reaction is a high molecular mass protein complex. In analogy to the ribosome and spliceosome it has been named the editosome (reviewed in Göringer, 2012b). Editosomes function as a catalytic platform for all steps of the reaction cycle and execute the processing reaction in a cascade of enzyme-driven steps. This includes endo- and exonucleases, a terminal uridylyltransferase (TUTase), RNA-ligases as well as RNA-chaperone activities.
The basal steps of the editing reaction cycle have been unraveled with the help of an in vitro RNA-editing assay (Seiwert and Stuart, 1994; Kable et al., 1996; Igo et al., 2000; Igo et al., 2002). The protocol represents a chemically simplified experimental system in which both, the substrate pre-edited mRNAs as well as the trans-acting gRNAs are mimicked by short RNA-oligonucleotides. Using purified editosome preparations, the pre-mRNA oligonucleotides are edited in a gRNA-dependent manner either by inserting 3 U-nt or deleting 4 U's. Importantly, in order to sidestep the rate-limiting endonucleolytic cleavage of the pre-mRNAs, the corresponding oligoribonucleotides are "pre-cleaved", i.e., they are provided as 5'- and 3'-pre-mRNA cleavage (Cl)-fragments. The 5'-Cl oligoribonucleotides are typically 5'-(32P)-labeled to enable a radioactive readout of the in vitro reaction.
Here we describe a new RNA-editing assay (Del Campo et al. 2020). The test system is a revised version of the established in vitro RNA-editing assays (Stuart et al., 1998), however, instead of using radioactively labeled substrate-RNAs it utilizes fluorophore-derivatized RNA-oligonucleotides. This allows the usage of automated, highly parallelized capillary electrophoresis (CE)-instruments coupled to laser-induced fluorescence (LIF) detection systems. It also enables multiplex-type fluorophore-labeling experiments to monitor multiple RNAs in one experiment. Furthermore, we designed new sets of pre-mRNA and gRNA-mimicking oligonucleotides. The different RNAs represent synthetic, non-natural sequences not found in the mitochondrial genomes of kinetoplastid organisms and they are, aside from the editing site, identical for both the U-insertion and the U-deletion reaction. This avoids potential sequence context issues and enables the side-by-side monitoring of both editing reactions in one reaction vial. Furthermore, we designed phosphorothioate (PS)-modified versions of the substrate RNAs. Phosphorothioate internucleotide linkages increase the RNase-stability of the pre-mRNA and gRNA-mimicking oligonucleotides, which permits the usage of editosome-enriched mitochondrial protein extracts instead of highly purified editosome preparations. Lastly, we optimized the work flow of the assay by adjusting the material quantities and reaction volume to multiwell-plate formats. The assay satisfies all signal-to-noise criteria for high-throughput screening methods (Del Campo et al., 2020) and is able to derive quantitative data for every step of the catalytic reaction cycle. As such the protocol should be helpful in the search for RNA-editing specific inhibitory compounds. The general workflow of the Fluorescence-based Insertion/Deletion Editing (FIDE)-assay is summarized in Figure 1.
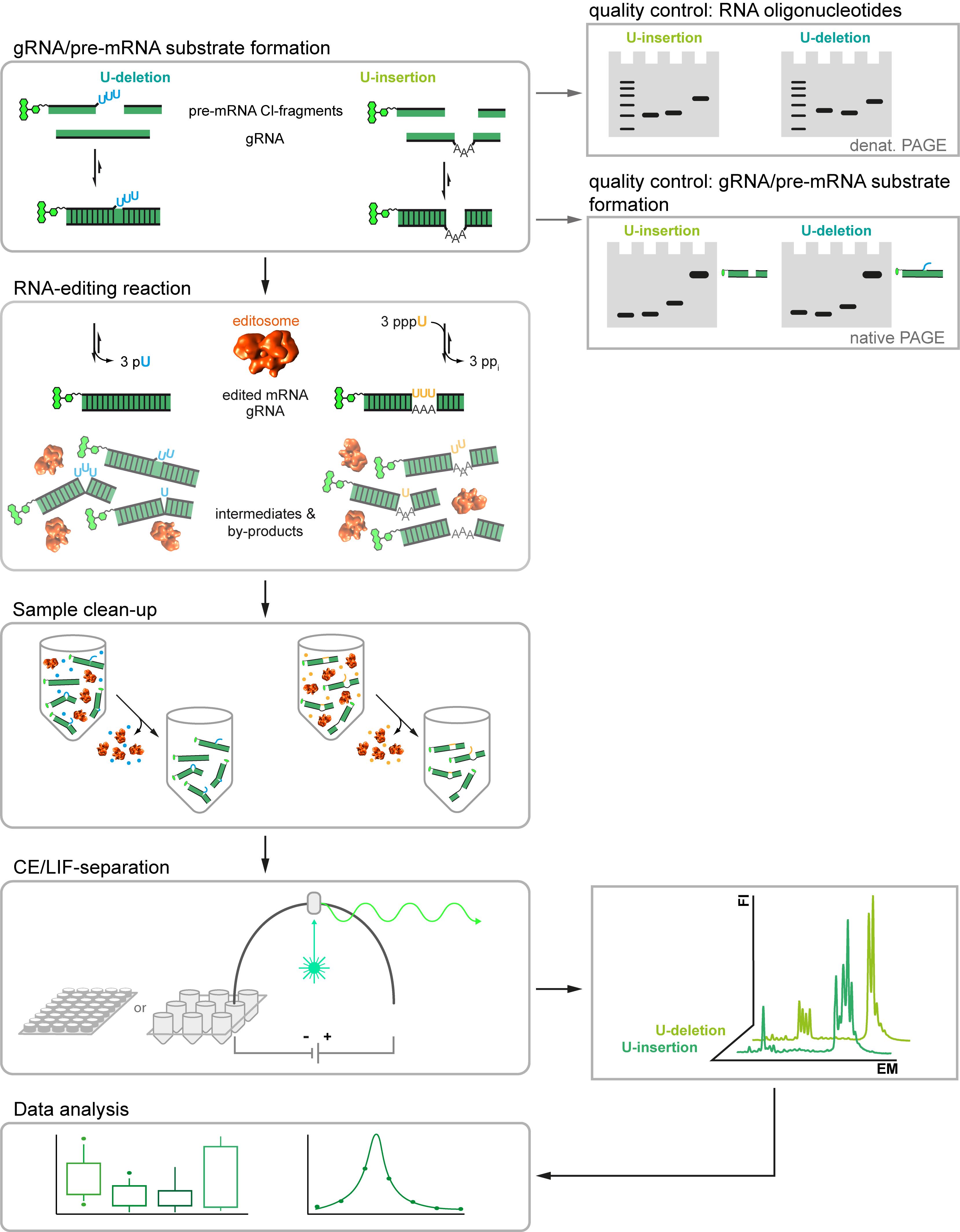
Figure 1. Workflow of the Fluorescence-based Insertion/Deletion Editing = FIDE-assay. Central steps of the protocol are on the left and boxed. Quality assessment steps and machine read-outs are illustrated on the right.
Materials and Reagents
All reagents, consumables and equipment are to be viewed as possible options. Unless stated otherwise, chemicals and consumables are stored at room temperature.
Solvents and Reagents
MeOH ≥99% p.a. (CARL ROTH, catalog number: 8388 )
EtOH ROTIPURAN® ≥99.8%, p.a. (CARL ROTH, catalog number: 9065 )
CHCl3 ≥99%, p.a (CARL ROTH, catalog number: 3313 )
Roti®-Phenol (TE-buffer equilibrated, pH 7.5-8.0) (CARL ROTH, catalog number: 0038 , store at 4 °C)
Isoamyl alcohol (3-Methyl-1-butanol), p.a. (Merck, catalog number: 100979 )
Glycerol ≥99.7%, p.a. (CARL ROTH, catalog number: 6962 , store at 4 °C)
Bradford assay reagent – detergent compatible (ThermoFisher Scientific, catalog number: 23246S , store at 4°C)
Phenol/Chloroform/Isoamylalcohol (PCI) (see Recipes, store at 4 °C)
Acids and Bases
Buffer Compounds and Salts
Tris-(hydroxymethyl)-amino methane (TRIS) PUFFERAN® (CARL ROTH, catalog number: 4855 )
N-2-Hydroxyethylpiperazine-N'-2-ethane sulphonic acid (HEPES) PUFFERAN® ≥99.5%, p.a. (CARL ROTH, catalog number: 9105 )
Imidazole PUFFERAN® ≥99%, p.a. (CARL ROTH, catalog number: X998 )
KCl (CARL ROTH, catalog number: 6781 )
MgCl2·6H2O (CARL ROTH, catalog number: 2189 )
NaCl (CARL ROTH, catalog number: 3957 )
Reducing and Chelating reagents
1,4-Dithiothreitol (DTT) ≥99%, p.a. (CARL ROTH, catalog number: 6908 , store at 4 °C)
Tris (2-carboxyethyl)phosphine hydrochloride (TCEP) ≥98% (Sigma-Aldrich, catalog number: C4706 , store at 4 °C)
Na2-ethylenediaminetetraacetate (Na2EDTA)·2H2O (CARL ROTH, catalog number: 8043 )
Ethylene glycol bis (β-aminoethylether) tetraacetic acid (EGTA) ≥99%, p.a. (CARL ROTH, catalog number: 3054 )
Ribonucleotides
Gel Electrophoresis Consumables
Acrylamide/Bisacrylamide (19:1) Rotiphorese®Gel 40 (CARL ROTH, catalog number: 3030 , store at 4 °C)
Urea (CARL ROTH, catalog number: 2317 )
Ammonium peroxydisulphate (CARL ROTH, catalog number: 9592 )
N,N,N',N'-Tetramethylethylendiamine (CARL ROTH, catalog number: 2367 )
Bromophenol Blue, Na+-salt (Merck, catalog number: 108122 )
Xylene cyanol FF, Na+-salt (Sigma-Aldrich, catalog number: X4126 )
Toluidine Blue O (Sigma-Aldrich, catalog number: T3260 )
6× Native PAGE gel-loading buffer (see Recipes)
2× Denaturing PAGE gel-loading buffer (see Recipes)
TBE buffer, pH 8.3 (see Recipes)
Gel staining solution (see Recipes)
Gel destaining solution (see Recipes)
Capillary Electrophoresis Consumables (see Note 1)
Glass capillary (Polymicro Technologies, catalog number: 1068150017 )
Performance optimized Polymer 6 (POP-6TM) (ThermoFisher Scientific, catalog number: 402837 , store at 4 °C)
10× CE/LIF-buffer (Sigma-Aldrich, catalog number: B4930 , store at 4 °C)
H2O LiChrosolv® (Merck Millipore, catalog number: 115333 )
Highly deionized formamide (Hi-DiTM Formamide) (ThermoFisher Scientific, catalog number: 4311320, store at -20 °C )
Editosomes and RNA-editing reaction mix
Affinity-purified editosomes in 10 mM HEPES/KOH, pH 7.5, 1 mM Mg(OAc)2, 75 mM NaCl, 0.1% (w/v) NP-40, 0.1 mM TCEP, 1 mM imidazole and 10 mM EGTA, 20% (v/v) glycerol (see Note 2, store at -20 °C)
Alternatively for HTS-applications: Editosome-enriched Trypanosoma brucei mitochondrial protein extracts in 20 mM HEPES/KOH, pH 7.5, 10 mM Mg(OAc)2, 30 mM KCl, 0.5 mM DTT, 20% (v/v) glycerol (see Note 2, store at -20 °C)
3x RNA-editing reaction mix (see Recipes, store at -20 °C)
Oligoribonucleotides (see Notes 3 and 4)
Standard-FIDE assay:
U-insertion
5’-FAM-synCl14ins: FAM-(CH2)6-AAAGGAAAUAUAGU
3'-synCl16: pAGGUGAUUCCAUUGAG-(CH2)6-NH2
syn-gRNAins: CUCAAUGGAAUCACCUAAAACUAUAUUUCCUUU
U-deletion
5'-FAM-synCl17del: FAM-(CH2)6-AAAGGAAAUAUAGUUUU
3'-synCl16: pAGGUGAUUCCAUUGAG-(CH2)6-NH2
syn-gRNAdel: CUCAAUGGAAUCACCUACUAUAUUUCCUUU
Standard-FIDE assay – phosphorothiate (PS)-modified (*) RNAs:
U-insertion
5’-FAM_modCl14ins: FAM-(CH2)6-AAAGGAAAU*A*U*A*GU
3’-modCl16: pAG*G*U*GAUUCCAUUGAG-(CH2)6-NH2
modgRNAins: C*U*C*AAUGGAAUCACCUAAAACUAUAUUUCC*U*U*U
U-deletion
5’-FAM_modCl17del: FAM-(CH2)6-AAAGGAAAU*A*U*A*GUUUU
3’-modCl16: pAG*G*U*GAUUCCAUUGAG-(CH2)6-NH2
modgRNAdel: C*U*C*AAUGGAAUCACCUACUAUAUUUCC*U*U*U
Multiplex ("one-pot") U-insertion/U-deletion assay:
U-insertion
5'-TAMRA_Cl18: TAMRA-(CH2)6-GGAAGUAUGAGACGUAGG
3'-Cl13: pAUUGGAGUUAUAG-(CH2)6-NH2
gRNAins: CUAUAACUCCGAUAAACCUACGUCUCAUACUUCC
U-deletion
5’-FAM_Cl22: FAM-(CH2)6-GGAAAGGGAAAGUUGUGAUUUU
3'-Cl15: pGCGAGUUAUAGAAUA-(CH2)6-NH2
gRNAdel: GGUUCUAUAACUCGCUCACAACUUUCCCUUUCC
Dual fluorophore assay:
U-insertion
5'-TAMRA_Cl18: TAMRA-(CH2)6-GGAAGUAUGAGACGUAGG
3'-Cl13_FAM: pAUUGGAGUUAUAG-(CH2)6-FAM
gRNAins: CUAUAACUCCGAUAAACCUACGUCUCAUACUUCC
U-deletion
5’-FAM_Cl22: FAM-(CH2)6-GGAAAGGGAAAGUUGUGAUUUU
3’-Cl15_TAMRA: pGCGAGUUAUAGAAUA-(CH2)6-TAMRA
gRNAdel: GGUUCUAUAACUCGCUCACAACUUUCCCUUUCC
RNA-size standards:
3'-FAM_13: AUUGGAGUUAUAG-(CH2)6-FAM
5'-FAM_22: FAM-(CH2)6-ggaaagggaaaguugugauuuu
5'-FAM_37: FAM-(CH2)6-ggaaagggaaaguugugauuuugcgaguuauagaaua
5’-FAM_44: FAM-(CH2)6-CUAGUACUCUCAUCAACAUAAGUCUCAUACUUCCGACAUGCA-CG
Reaction vials:
Equipment
Spectrophotometer (Thermo Scientific, model: NanoDrop 2000C )
Thermocycler (Biometra, T3 )
CE/LIF-based DNA-sequencer (Applied Biosystems, model: ABI PRISM 310 Genetic analyzer )
Benchtop Centrifuge (Eppendorf, model: 5417 R )
SpeedVac Vacuum Concentrator (Savant Instruments, model: SVC-100 )
Dry-Block Heating System (Grant, model: QBD2 )
Vortex Mixer (Scientific Instruments, model: Vortex Genie 2 )
DC-High Voltage Power Supply (Biometra, model: P30 )
Gel-Electrophoresis Chamber (BIO-RAD, model: Mini-PROTEAN® Tetra Cell)
Micropipettes (Gilson, models: Pipetman P10, P20, P200, P1000 )
Gel Documentation System (INTAS, model: Gel iX20 Imager )
Software
R (The R Project for Statistical Computing, https://www.r-project.org/)
Programmer-type text editor (such as Vim, https://www.vim.org or Atom, https://atom.io)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Leeder, M., Kruse, E. and Göringer, H. U. (2021). Trypanosomatid, fluorescence-based in vitro U-insertion/U-deletion RNA-editing (FIDE). Bio-protocol 11(5): e3935. DOI: 10.21769/BioProtoc.3935.
分类
分子生物学 > RNA
系统生物学 > 转录组学
生物化学 > RNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










