- EN - English
- CN - 中文
Molecular and Phenotypic Characterization Following RNAi Mediated Knockdown in Drosophila
RNAi介导的基因敲降后果蝇的分子和表型特征
(*contributed equally to this work) 发布: 2021年02月20日第11卷第4期 DOI: 10.21769/BioProtoc.3924 浏览次数: 5632
评审: Anonymous reviewer(s)
Abstract
Loss of function studies shed significant light on the involvement of a gene or gene product in different cellular processes. Short hairpin RNA (shRNA) mediated RNA interference (RNAi) is a classical yet straightforward technique frequently used to knock down a gene for assessing its function. Similar perturbations in gene expression can be achieved by siRNA, microRNA, or CRISPR-Cas9 methods also. In Drosophila genetics, the UAS-GAL4 system is utilized to express RNAi and make ubiquitous and tissue-specific knockdowns possible. The UAS-GAL4 system borrows genetic components of S. cerevisiae, hence rule out the possibility of accidental expression of the system. In particular, this technique uses a target-specific shRNA, and the expression of the same is governed by the upstream activating sequence (UAS). Controlled expression of GAL4, regulated by specific promoters, can drive the interfering RNA expression ubiquitously or in a tissue-specific manner. The knockdown efficiency is measured by RNA isolation and semiquantitative RT-PCR reaction followed by agarose gel electrophoresis. We have employed immunostaining procedure also to assess knockdown efficiency.
RNAi provides researchers with an option to decrease the gene product levels (equivalent to hypomorph condition) and study the outcomes. UAS-GAL4 based RNAi method provides spatio-temporal regulation of gene expression and helps deduce the function of a gene required during early developmental stages also.
Background
Drosophila melanogaster (fruit fly) is a versatile model organism frequently used in research laboratories. Fruit flies are easy to handle, propagate, and maintain. Moreover, the elaborate yet short life span with high fecundity is an added advantage of Drosophila. The facile nature of Drosophila genetics tools helps develop a comprehensive understanding of a gene function. Since 60% of the Drosophila genes are homologous to human genes, and other advantages mentioned earlier, Drosophila is an obvious model organism of choice to study in vivo gene functions.
The UAS-GAL4 system was used for in vivo activation of transcription of genes. Moreover, it was suggested that antisense RNAs could be expressed using the UAS-GAL4 system to achieve significant inhibition of gene expression (Brand and Perrimon, 1993). For using the UAS-GAL4 system in Drosophila, transgenic fly lines carrying the upstream activating sequence (UAS) capable of regulating RNAi expression are generated. Genome-scale shRNA-dependent RNAi resource was generated for stage-specific effective knockdown of Drosophila genes (Ni et al., 2011). RNAi lines are publicly available from Vienna Drosophila Resource Center (VDRC, maintaining GD and KK RNAi lines) and Drosophila RNAi Screening Center (TRiP and shRNA lines were generated at DRSC and are currently maintained and available from Bloomington Drosophila Stock Center, BDSC). The detailed methodology for the generation of different RNAi lines can be found at the VDRC stock center (https://stockcenter.vdrc.at/control/main) and DRSC (https://fgr.hms.harvard.edu/) websites.
When the RNAi fly line is crossed with the GAL4 driver fly line, the progenies can have both components of the UAS-GAL4 system expressed. The upstream promoter guides GAL4 expression, and wherever the GAL4 is expressed, it binds to the UAS sequence with high affinity, allowing expression of the RNAi component (Figure 1). In the absence of GAL4 expression, the transgenic RNAi flies behave like wild-type due to lack of dsRNA expression. This method allows the knockdown of the desired gene in a spatiotemporal manner. Classically, X-ray mutagenesis, and P-element mobilization have been used to achieve the loss of gene expression. Further, the EMS mutagenesis screens have been utilized to induce point mutation, occasionally leading to truncations affecting gene expression. The RNAi mediated knockdown is rather facile and less labor-intensive.
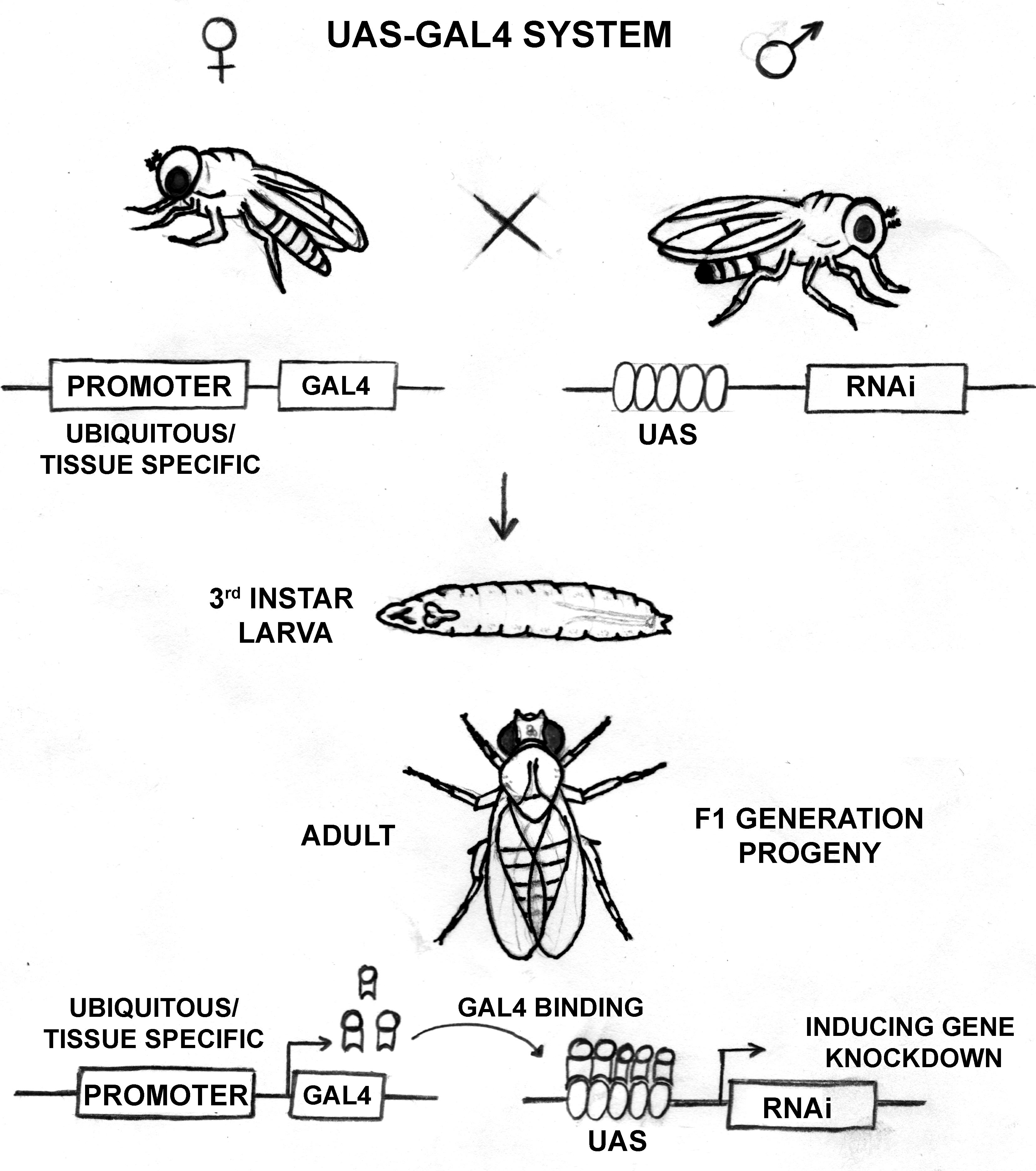
Figure 1. Schematic diagram detailing the functioning of the UAS-GAL4 system in Drosophila Due to varying GAL4 expression degrees in different tissues, a gradient in the knockdown can be seen in different tissues. Besides, temperature-dependent regulation on the GAL4 function provides an advantage of regulating knockdown levels. Together, these handles on gene expression regulation using the UAS-GAL4 system provide more information regarding the functions of our gene of interest. Importantly, this method provides a controlled spatiotemporal regulation on RNAi mediated knockdown and can help study functions of developmentally essential genes, genes with housekeeping functions, and mutations in genes associated with harmful consequences. This method also helps in the functional characterization of a new gene like dElys (Mehta et al., 2020).
A typical qRT-PCR or semiquantitative PCR validates the reduction in gene expression levels upon RNAi mediated knockdown. The knockdown efficiencies can be further assessed by detecting the desired gene product levels by immunostaining and Western blotting methods. We have utilized the RNAi mediated knockdown in salivary glands and assessed its effectiveness using immunostaining or assessing mRNA levels by semiquantitative PCR. We successfully depleted nucleoporins in eyes and wings using the RNAi-mediated knockdown method and reported the importance of nucleoporins in tissue and organism development.
Materials and Reagents
1.5 ml microcentrifuge tubes (Genaxy, catalog number: GEN-MT-150-C-S )
Barrier tips
0.5-10 µl tips (Axygen Scientific, catalog number: TF-300-L-R-S )
1-20 µl tips (Axygen Scientific, catalog number: TF-20-L-R-S )
1-200 µl tips (Axygen Scientific, catalog number: TF-200-L-R-S )
100-1,000 µl tips (Axygen Scientific, catalog number: TF-1000-L-R-S )
Glass vials and bottles
SYLGARD 184 Silicone elastomer (DOW, catalog number: 1673921 )
Micropipette tips (2, 20, 200, 1,000 µl)
0.5-10 µl tips (Axygen Scientific, catalog number: T-300-L-R-S )
1-200 µl tips (Axygen Scientific, catalog number: TR-222-C-L-R-S )
100-1,000 µl tips (Axygen Scientific, catalog number: T-1000-C-L-R-S )
3 cm dish (Eppendorf, catalog number: 00 30700112 )
Food bottles
CO2 pads
Cotton plugs
Corn flour
Table sugar
Yeast extract (HiMedia, catalog number: RM027 )
Agar (Merck, catalog number: A5306 )
Dextrose (HiMedia, catalog number: GRM077 )
Methyl-4-Hydroxybenzoate (Sigma-Aldrich, catalog number: H5501 )
Ortho-Phosphoric Acid (Merck, catalog number: 100573 )
Propionic Acid (HiMedia, catalog number: GRM3658 )
Ethanol (Changshu Hongsheng Fine Chemical CHN01 )
Liquid Nitrogen
RNA isolation kit (Favorgen, catalog number: FATRK001 )
RNaseZAP (Thermo Scientific, catalog number: AM9780 )
DNase (Thermo Scientific, catalog number: EN0523 )
Diethyl pyrocarbonate (HiMedia, catalog number: MB076 )
Ethanol (MP Biomedicals, catalog number: 180077 )
Formamide (Sigma-Aldrich, catalog number: F7503 )
6× DNA loading dye purple (New England Biolabs, catalog number: B7024S )
cDNA synthesis kit (iScript, Bio-Rad, catalog number: 1708891 )
Agarose (Invitrogen, catalog number: 16500500 )
Ethidium bromide (EtBr) (MP Biomedicals, catalog number: 193993 )
Custom Primers (Integrated DNA Technology)
G9-Taq DNA polymerase (GCC Biotech, catalog number: G7115A )
dNTPs solution mix (New England Biolabs, catalog number: N0447L )
Sodium Chloride (Sigma-Aldrich, catalog number: S3014 )
Potassium Chloride (Sigma-Aldrich, catalog number: P5405 )
di-Sodium hydrogen phosphate heptahydrate (Merck, catalog number: 1065751000 )
Potassium dihydrogen phosphate (Sigma-Aldrich, catalog number: P9791 )
Tris-base (Sigma-Aldrich, catalog number: T6066 )
Glacial acetic acid (Merck EMPLURA, catalog number: 1.93402.2521 )
Carbon conductive tape (TED PELLLA Inc. catalog number: 16073-1 )
Aluminum stub (TED PELLLA Inc. catalog number: 16111-9 )
Glutaraldehyde (Sigma-Aldrich, catalog number: G5882 )
Gold target (Quorum Technology inc, Catalog number: SC502-314A )
EDTA (Merck EMPARTA, catalog number: 1.93312.1021 )
Paraformaldehyde (Sigma-Aldrich, catalog number: 158127 )
Triton X-100 (Sigma-Aldrich, catalog number: X100 )
Glass Slide (Borosil, catalog number: 9100P02 )
Coverslips (Borosil, catalog number: 9115S01 )
Bovine serum albumin (Merck, catalog number: 621650500501730 )
Fluoroshield with DAPI (Sigma-Aldrich, catalog number: F6057 )
mAb414 (Biolegend 902901) - mAb414 recognizes FG-repeat rich four distinct nucleoporins of nuclear pores
Anti-dElys (Mehta et al, 2020)
Alexa Fluor-488 (Invitrogen, catalog number: A-11029 )
Alexa Fluor-568 (Invitrogen, catalog number: A-11036 )
Coverslip sealant (Transparent nail polish)
Diethyl ether (Merck, catalog number: 107026 )
Phosphate Buffer Saline (1×) (see Recipes)
PBS-T (see Recipes)
Fly food ingredients (see Recipes)
RNA loading dye (see Recipes)
DEPC treated water (see Recipes)
TAE Buffer (50×) (see Recipes)
4% Paraformaldehyde (see Recipes)
Control Wild type (w1118)
Driver lines (in house generated combinations)
+/+; Actin5C-GAL4/CyO-GFP; UAS-Dicer/UAS-Dicer
+/+; wingless-GAL4/wingless-GAL4; UAS-Dicer/UAS-Dicer
+/+; eyeless-GAL4/eyeless-GAL4; UAS-Dicer/UAS-Dicer
+/+; v103547/v103547; UAS-Dicer/UAS-Dicer (dElys RNAi KK line combined with UAS-Dicer, in house generation)
dElys RNAi KK line (VDRC, VDRC ID: 103547)
Nup160 RNAi GD line (VDRC, VDRC ID: 21937)
Sec13 RNAi KK line (VDRC, VDRC ID: 110428)
Nup107 RNAi GD line (VDRC, VDRC ID: 22407)
Equipment
Micropipette (Nichiriyo, Gilson, Corning)
BOD Incubators
Millipore water purification unit
Genova Nanodrop (Bibby Scientific, model: 737501 )
Forceps Dumont 5 (Fine Science Tools, catalog number: 11295-10 )
Forceps Dumont 55 (Fine Science Tools, catalog number: 11295-51 )
Scissors (Fine Science Tools, catalog number: 91500-09 )
Leica DM2500
Fluorescent stereomicroscope (Leica, model: M205 FA )
Confocal Laser Scanning Microscope (Zeiss, model: LSM780 )
Stereomicroscope (Leica, model: S6E )
Scanning Electron microscope (Zeiss, model: Gemini II Ultra plus )<
Tabletop centrifuge (Eppendorf, model: 5424, Thermo Scientific MicroCL 21R )
PCR machine (Applied Biosystems, model: 2720 Thermal cycler )
Water bath (Grant-Bio, model: PSU-10i )
Gel running apparatus (CBS Scientific, model: GCMGU-202T )
Power packs (CBS Scientific, model: EPS 300 )
UVP MultiDoc-It Digital Imaging System (UVP, catalog number: 97-0192-02 )
Software
ZEN 3.2 Blue edition (Zeiss)
ImageJ/Fiji (Free, NIH)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mehta, S. J. K., Joshi, P. A. and Mishra, R. K. (2021). Molecular and Phenotypic Characterization Following RNAi Mediated Knockdown in Drosophila. Bio-protocol 11(4): e3924. DOI: 10.21769/BioProtoc.3924.
- Mehta, S. J. K., Kumar, V. and Mishra, R. K. (2020). Drosophila ELYS regulates Dorsal dynamics during development. J Biol Chem 295(8): 2421-2437.
分类
发育生物学 > 细胞信号传导 > 细胞凋亡
细胞生物学 > 细胞染色 > 蛋白质
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link