- EN - English
- CN - 中文
Bacterial Conjugation Protocol for Ruminant Mycoplasmas
反刍动物支原体细菌接合的实验方法
(*contributed equally to this work) 发布: 2021年01月20日第11卷第2期 DOI: 10.21769/BioProtoc.3893 浏览次数: 3433
评审: Alba BlesaOscar Quijada Pich Alvaro Eduardo Galvis
Abstract
In Mycoplasma agalactiae, two simultaneous processes of DNA transfer have been described that require direct cell-to-cell contact and are similar to conjugation. One involves the self-transmission of an integrative conjugative element (ICE) while the second concerns the horizontal transfer of large and small fragments of chromosomal DNA. Here, we describe an optimized conjugation protocol for the horizontal transfer of ICE or chromosomal DNA carrying antibiotic resistance markers (i.e., tetracycline, gentamicin, puromycin) from donor to recipient mycoplasma cells. Calculation of the conjugation frequencies, selection and characterization of transconjugants are detailed. This protocol has been developed with M. agalactiae but has been successfully used for M. bovis and can be adapted to other related mycoplasma species.
Keywords: Mycoplasma (支原菌)Background
Conjugative, horizontal DNA transfer is a key player of microbial diversification. By promoting an active transfer of DNA through close cell-to-cell contact (Lederberg and Tatum, 1946), this phenomenon facilitates the rapid acquisition of new traits from external resources. Mycoplasmas (class Mollicutes) are wall-less bacteria with reduced genomes whose evolution has been considered to be only driven by gene loss and in which horizontal gene transfer (HGT) was long thought to be marginal. During this past decade, comparative genomic analyses have revisited this paradigm and have shown (i) the occurrence of HGTs events in between mycoplasma species sharing same hosts (Sirand-Pugnet et al., 2007) and (ii) the presence of integrative and conjugative elements (ICEs) in a large number of sequenced mycoplasma genomes (Calcutt et al., 2002; Marenda et al., 2006; Dordet-Frisoni et al., 2013; Tardy et al., 2015; Meygret et al., 2019), raising the prospect that these minimal bacteria might be able to conjugate.
In vitro conjugation-like process has been demonstrated in Mycoplasma agalactiae (Dordet-Frisoni et al., 2013, 2014 and 2019), a mycoplasma species pathogenic for ruminants. Mating assays were based on mixing in liquid culture two strains tagged with antibiotic resistant genes and resulted in a bidirectional DNA transfer process (Dordet-Frisoni et al., 2013 and 2014). The first is the conventional horizontal dissemination of a mycoplasma ICE (MICE), from ICE-positive to ICE-negative cells (Dordet-Frisoni et al., 2013). The second concerns the transfer of large and small blocks of chromosomal DNA from ICE-negative to ICE-positive cells and was further referred to as MCT for mycoplasma chromosomal transfer (Dordet-Frisoni et al., 2014 and 2019; Faucher et al., 2019). M. agalactiae conjugation has been shown to rely on the presence of an ICE in at least one parent cell, but both DNA transfers are physically independent (taking place in opposite direction) and occur at the same frequency (around 10-8 transconjugants/total CFU) (Dordet-Frisoni et al., 2014).
The detailed protocol presented here describes an optimized mating procedure developed for the conjugation of M. agalactiae and related species. For instance, as described in Dordet-Frisoni et al. (2014), this protocol has also been successfully applied to M. bovis and can most likely be adapted to other mycoplasma species with some adjustments.
Materials and Reagents
Sterile tips for micropipettes (STARLAB, catalog numbers: S1122-1830 [1,000 µl], S1120-3810 [10-20 µl] )
Sterile 1.5 ml and 2 ml microcentrifuge tubes (SORENSON, catalog number: 017040A; TREFF, catalog number: 96.09329.9.01)
Sterile plastic Petri dish plates (90 mm diameter, Clearline, catalog number: 076084BS )
0.22 µm pore filter (PVDF 33 mm, ClearLine, Dutscher, catalog number: 051733)
Donor and recipient cells, harbouring different selective markers for antibiotic selection #
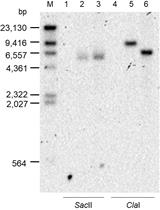
#Note: M agalactiae strains* were tagged with selectable markers previously introduced by transposition using a pMT85-derived plasmid (Figure 1) (Zimmerman and Herrmann, 2005). pMT85 and derivatives bear a mini-Tn with either the aacA-aphD gentamicin resistance cassette, the tetM determinant for resistance to tetracycline or the pac gene conferring puromycin resistance. Marker genes are flanked by two Tn4001 inverted repeats (IRs). Since the mini-Tn backbone contains no transposase gene (it is located elsewhere in the pMT85), its insertion into the cell chromosome is stable (Baranowski et al., 2010).
Several Mycoplasma agalactiae strains such as 5632, PG2, 4867, 14628 and 4055, have been successfully employed in mating assays (Dordet-Frisoni et al., 2019), but conjugation experiment has been also successful with other ruminant mycoplasmas species such as M. bovis PG45 strain (Dordet-Frisoni et al., 2014) and M. mycoides subsp. capri GM12 strain (personal data). For conjugation to occur, at least one of the two partners should carry an ICE. “Concerning chromosomal transfers (MCT), only two M. agalactiae strains have been documented as recipient cells, namely 5632 and 4867, while all strains tested so far can act as donor. As for ICEA, all strains carrying an active ICE are potentially donor cell. All parental M. agalactiae strains cited above can be acquired at the Vigimyc collection (Chazel et al., 2010).
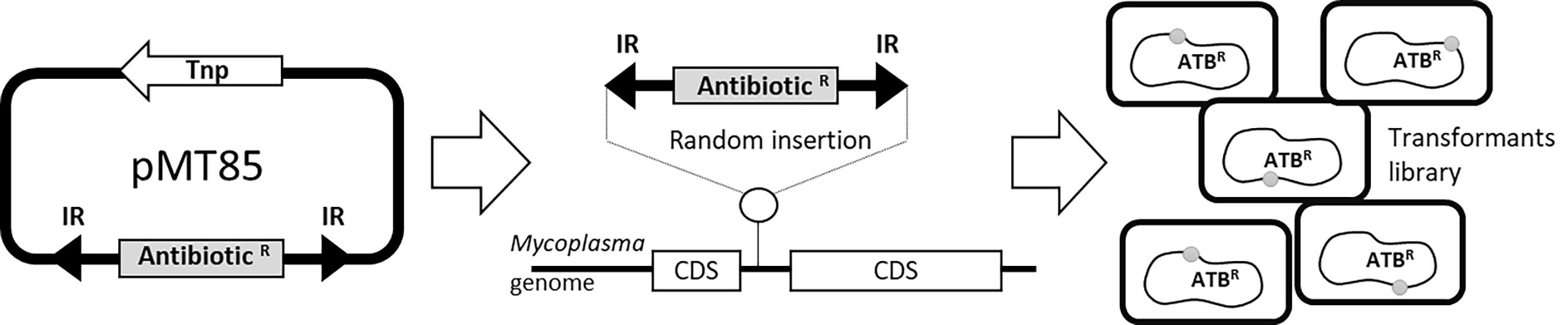
Figure 1. Schematic representing M. agalactiae antibiotic tagging procedure. Selectable markers are introduced by transposition using the mini transposon pMT85, which bear antibiotic resistance genes as selection markers (AntibioticR). The AntibioticR marker is flanked by two inverted (IR) Tn4001 repeats. During insertion, the transposase (Tnp) which is located outside the IRs is eliminated and the selection marker is integrated into the chromosome in a random (in coding (CDS) or non-coding sequences) and a stable manner. This experiment results in mycoplasmas transformants tagged with resistance markers (ATBR) at different locations on the chromosome.Appropriate selectable antibiotics:
Gentamicin (GmR) (Gibco, catalog number: 15750037 )
Tetracycline (TetR) (Sigma-Aldrich, catalog number: T3383 )
Puromycin (PurR) (Fisher, catalog number: 10054207 )
Ampicillin (Sigma-Aldrich, catalog number: A9518 )
PPLO broth (Difco, catalog number: 255420 )
Tryptone peptone (Difco, catalog number: 211705 )
Bacto peptone (Difco, catalog number: 211677 )
FBS (Fetal Bovine Serum) (Gibco, catalog number: 10270-106 )
CMRL 1066 (10x) (Gibco, catalog number: 21540-026 )
TC yeastolate (Difco, catalog number: 255772 )
Yeast extract 15% (Gibco, catalog number: 18180-059 )
D-(+)-Glucose (Sigma-Aldrich, catalog number: G6152 )
Phenol red 0.5% (Sigma-Aldrich, catalog number: P0290 )
Na Pyruvate (Sigma-Aldrich, catalog number: P2256 )
Select agar (Invitrogen, catalog number: 30391-023 )
NaOH (Sigma-Aldrich, catalog number: S8045 )
D-PBS (Gibco, catalog number: 14190169 )
Tween-20 (Sigma-Aldrich, catalog number: P9416 )
Ultra pure Tris (Invitrogen, catalog number: 15504020 )
HCl (Sigma-Aldrich, catalog number: 320331 )
Proteinase K 20 mg/ml (Qiagen catalog number: 19131 )
Taq DNA Polymerase (New England Biolabs catalog number: M0267L )
dNTP mix 10 mM (New England Biolabs catalog number: N0447S )
Taq Buffer 10x (New England Biolabs catalog number: M0267L )
Sterile SP4 liquid medium (see Recipes)
Sterile SP4 agar plates (see Recipes)
M. agalactiae Lysis buffer (see Recipes)
Equipment
Air-flow cabinet (Faster, model: SF Classic 212 )
37 °C incubator (Firlabo, model: FA Type B 407L-FA407B )
Microcentrifuge with rotor for 1.5-2 ml tubes (Eppendorf, model: 5424 )
Vortex (Scientific Industries, model: GENIE 2 )
Binocular magnifier (Olympus, model: SZ 60 )
Multiblock heater (Lab Line Instruments, model: 2052-1CCE )
Thermocycler (Mastercycler EP Eppendorf Gradient, model: 5341 )
Micropipettes (GIilson Pipetman Classic 10 μl to 1 ml range, Dutscher, catalog numbers: 066002 , 066003 , 066004 , 066005 and 066006 )
Graduated pipettes (Falcon 5 to 25 ml range pipettes, Dutscher, catalog numbers: 357543 , 357551 and 357535 )
Pipettor (Pipetboy Acu 2, INTEGRA, DUTSCHER, catalog number: 060201 )
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sagné, E., Citti, C. and Dordet-Frisoni, E. (2021). Bacterial Conjugation Protocol for Ruminant Mycoplasmas . Bio-protocol 11(2): e3893. DOI: 10.21769/BioProtoc.3893.
分类
微生物学 > 微生物遗传学 > 转化
微生物学 > 微生物生理学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link