- EN - English
- CN - 中文
Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) into Osteoclasts
人诱导多能干细胞分化破骨细胞的研究
发布: 2020年12月20日第10卷第24期 DOI: 10.21769/BioProtoc.3854 浏览次数: 5313
评审: Giusy TornilloAnthony FlamierSébastien Gillotin

相关实验方案
人 iPSC 衍生神经元与少突胶质细胞共培养用于髓鞘形成的小分子筛选分析
Stefanie Elke Chie [...] Maria Consolata Miletta
2025年05月05日 3362 阅读
Abstract
Defects in bone resorption by osteoclasts result in numerous rare genetic bone disorders as well as in some common diseases such as osteoporosis or osteopetrosis. The use of hiPSC-differentiated osteoclasts opens new avenues in this research field by providing an unlimited cell source and overcoming obstacles such as unavailability of human specimens and suitable animal models. Generation of hiPSCs is well established but efficient differentiation of hiPSCs into osteoclasts has been challenging. Published hiPSC-osteoclast differentiation protocols use a hiPSC-OP9 co-culture system or hiPSC-derived embryoid bodies (EBs) with multiple cytokines. Our three-stage protocol consists of 1) EB mesoderm differentiation, 2) expansion of myelomonocytic cells and 3) maturation of hiPSC-osteoclasts. We generate uniformly-sized EBs by culturing Accutase-dissociated hiPSCs on Nunclon Sphera microplates and promote EB mesoderm differentiation in a cytokine cocktail for 4 days. For Stage 2, EBs are transferred to gelatin-coated plates and cultured with hM-CSF and hIL-3 to expand the myelomonocytic population. By supplementing with vitamin D, hTGFβ, hM-CSF and hRANKL, cells collected at the end of Stage 2 are differentiated into mature osteoclasts (Stage 3). Compared to other techniques, our protocol does not require a co-culture system; induces EBs into mesoderm differentiation in a homogenous manner; uses less cytokines for differentiation; requires only a short time for osteoclast maturation and produces sufficient numbers of osteoclasts for subsequent molecular analyses.
Graphic abstract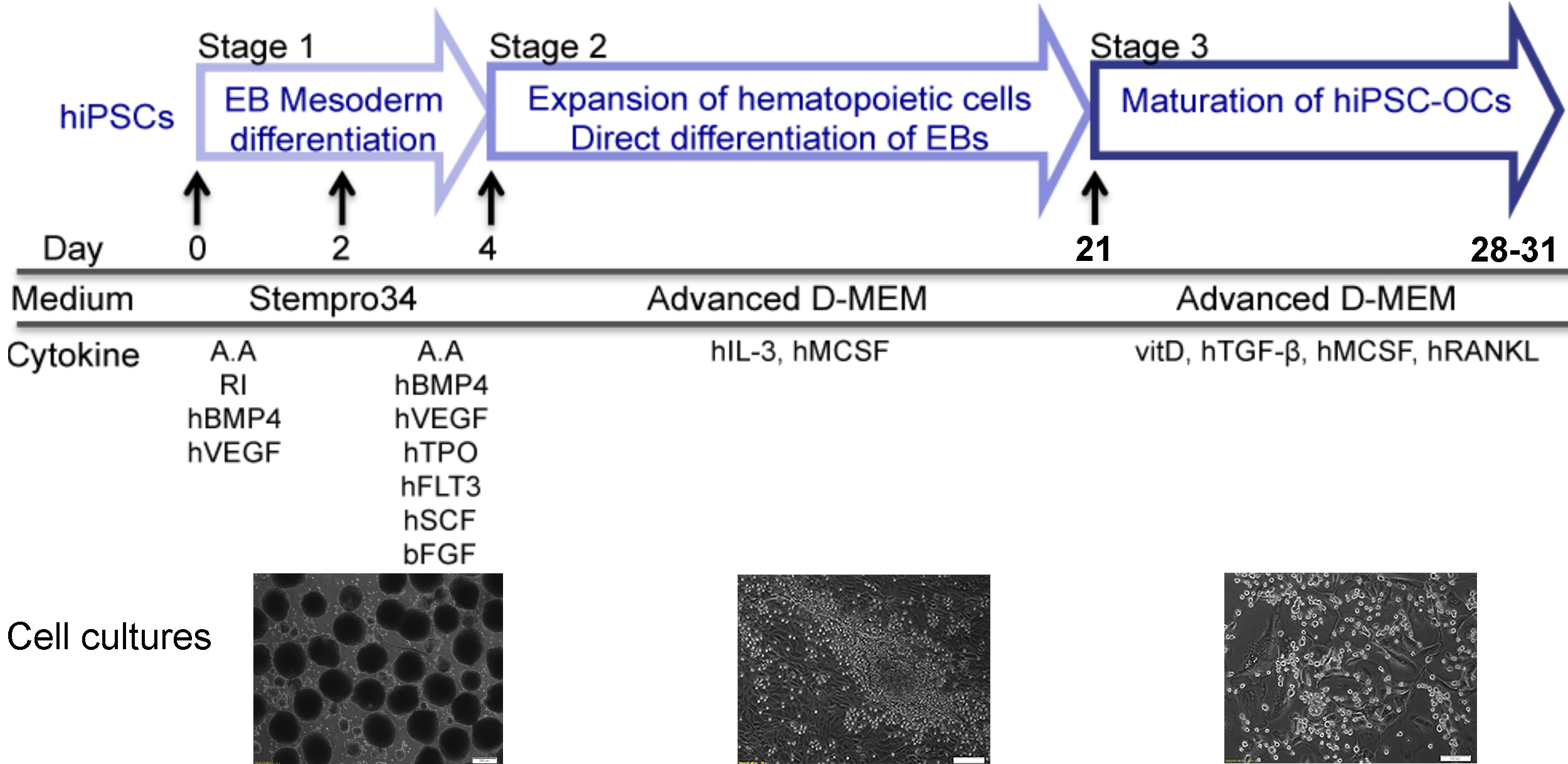
Background
The technology for generating patient-specific hiPSCs, which theoretically can be differentiated into any cell type, opens new avenues for medical research in disease modeling, including bone disorders (Deyle et al., 2012; Quarto et al., 2012a and 2012b; Cherry and Daley, 2013; Ding et al., 2013; Matsumoto et al., 2013; Chen et al., 2017). Research focusing on rare genetic bone disorders not only has the potential to find treatment for patients, but also contributes to a better understanding of skeletal biology. Many skeletal diseases involve dysfunctional osteoclasts, osteoblasts and/or osteocytes (Chen, 2014). Osteoclasts are bone resorbing cells and osteoblasts are bone forming cells. Osteocytes are derived from mature osteoblasts and are entrapped in the bone matrix that they produce (Bonewald, 2011). Bone marrow stromal cells (mesenchymal osteoblast-like cells) can be cultured from bone marrow, bone biopsies or bone excised during surgical procedures. Mesenchymal cells are proliferative and can be differentiated and passaged. The myelomonocytic population in bone marrow and peripheral blood can be differentiated into osteoclasts by culturing with human macrophage stimulating factor (M-CSF) and receptor activator of nuclear factor-kB ligand (RANKL) (Chen et al., 2011). However, once differentiated, these cells are terminally differentiated and can be used for experiments only once.
While reliable and consistent methods for reprogramming somatic cells into hiPSCs are well-established (Takahashi et al., 2007; Yu et al., 2007; Park et al., 2008; Stadtfeld et al., 2008; Sommer et al., 2009; Yu et al., 2009; Warren et al., 2010), differentiation of hiPSCs into bone cells is still more challenging. Several studies describe protocols for hiPSC differentiation into osteoblasts (Kanke et al., 2014; Kuhn et al., 2014; Ochiai-Shino et al., 2014; Kang et al., 2016), but there are relatively few protocols described for hiPSC differentiation into osteoclasts (Choi et al., 2009; Grigoriadis et al., 2010; Jeon et al., 2016). Choi et al. and Grigoriadis et al. differentiated hiPSC into osteoclasts via a hiPSC-OP9 co-culture system and through EB formation steps, respectively (Choi et al., 2009; Grigoriadis et al., 2010). A critical step for co-culture systems is to match cell densities of undifferentiated hiPSCs and OP9 cells, which can otherwise contribute to inconsistent outcomes. Generating EBs by conventional methods in cell culture dishes may cause variable EB sizes and can affect the differentiation efficiency. In addition, the use of complex cytokine cocktails is less economical. Jeon et al. (2016) reported a hiPSC-osteoblast and hiPSC-osteoclast co-culture system, which is less useful for investigating cell-autonomous osteoblast and osteoclast phenotypes in disease models.
Our protocol described here generates uniformly-sized EBs by dissociating hiPSC colonies into single cells and plating a fixed number of cells onto Nunclon Sphera microplates. EBs are stimulated to enter the mesoderm lineage in the first 4 days of differentiation and are transferred to gelatin-coated plates to expand the myelomonocytic population, which contains the osteoclast progenitors. Differentiation into osteoclasts is achieved by culturing these cells in the presence of vitamin D, hTGFβ, hM-CSF and hRANKL. The resulting mature and functional osteoclasts are tartrate-resistant acid phosphatase (TRAP)-positive multinucleated cells and are able to resorb bone.
Materials and Reagents
Materials
Pipet tips (200 μl, 1 ml)
Cell strainer 70 μm (Corning, catalog number: 352350 )
Nunclon Sphera 96-well plates, U-bottom (Thermo Fisher Scientific, catalog number: 174929 )
Tissue culture 96-well plate, polystyrene (Greiner Bio-One, catalog number: 655180 )
Tissue culture 6-well plate, polystyrene (BD, Falcon, catalog number: 08-772-1B )
Serological pipet, 5 ml (Fisher Scientific, catalog number: 13-678-11D )
Serological pipet, 10 ml (Fisher Scientific, catalog number: 13-678-11E )
15 ml polypropylene conical tubes (Thermo Scientific, catalog number: 339650 )
50 ml polypropylene conical tubes (Thermo Scientific, catalog number: 339652 )
Nalgene filter (0.2 μm PES membrane) 150 ml (Thermo Scientific, catalog number: 5650020 )
Nalgene filter (0.2 μm PES membrane) 250 ml (Thermo Scientific, catalog number: 5680020 )
Microcentrifuge tube (Fisher Scientific, catalog number: 05-408-129 )
hiPSCs generated from healthy donors (Chen et al., 2013 and 2017)
Matrigel (Corning, catalog number: 354234 )
PeproGrow embryonic stem cell (hESC) medium (Peprotech, catalog number: BM-hESC )
Accutase (Millipore Sigma, catalog number: SCR005 )
Phosphate buffered saline, no calcium, no magnesium (Thermo Fisher Scientific, catalog number: 10010049 )
DMEM/F12 medium (Thermo Fisher Scientific, catalog number: 11330032 )
Stempro-34 medium (Thermo Fisher Scientific, catalog number: 10639011 )
L-Glutamine (200 mM) (Thermo Fisher Scientific, catalog number: 25030081 )
1-Thioglycerol (MTG) (Sigma-Aldrich, catalog number: M6145 )
Ascorbic acid (Sigma-Aldrich, catalog number: A4034 )
MEM Non-essential amino acids (NEAA 100x) (Thermo Fisher Scientific, catalog number: 11140-050 )
Rock inhibitor Y-27632 (Selleck Chemicals, catalog number: S1049 )
Recombinant human BMP4 (Peprotech, catalog number: 120-05ET )
Recombinant human VEGF (Peprotech, catalog number: 100-20 )
Recombinant human TPO (Peprotech, catalog number: 300-18 )
Recombinant human Flt3-ligand (Peprotech, catalog number: 300-19 )
Recombinant human IL-3 (Peprotech, catalog number: 200-03 )
Recombinant human MCSF (Peprotech, catalog number: 300-25 )
Recombinant human RANKL (Peprotech, catalog number: 310-01 )
Recombinant human TGF-β1 (Peprotech, catalog number: 100-21C )
Recombinant human SCF (Peprotech, catalog number: 300-07 )
Recombinant human FGF basic (Peprotech, catalog number: 100-18B )
Gelatin 0.1% solution (EMD Millipore, catalog number: ES-006-B )
Advanced DMEM medium (Thermo Fisher Scientific, catalog number: 12491023 )
Alpha MEM medium (Thermo Fisher Scientific, catalog number: 12571063 )
Fetal bovine serum (Gibco, catalog number: 10437020 )
1α, 25-dihydroxyvitamin D3 (Sigma Aldrich, catalog number: D1530 )
CD14 antibody (FITC-conjugated; Biolegend, catalog number: 325604 )
CD43 antibody (APC-conjugated, Miltenyi Biotec, catalog number: 560198 )
CD45 antibody (APC-conjugated, Biolegend, catalog number: 304012 )
EDTA 0.5 M, pH 8.0 (Thermo Fisher Scientific, catalog number: 15575-038 )
TRIzol (Thermo Fisher Scientific, catalog number: 10296028 )
Direct-zol RNA (Zymo Research, catalog number: R2052 )
DNase I (Invitrogen, catalog number: 18068015 )
Superscript II reverse transcriptase (Invitrogen, catalog number: 18064071 )
SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, catalog number: 1725271 )
Glutaraldehyde 25% solution (Alfa Aesar, catalog number: A17876 )
Acid phosphatase, leukocyte (TRAP) kit (Sigma-Aldrich, catalog number: 387A )
Primers for detection of mesoderm differentiation and osteoclast maturation by qPCR (Table 1)
Matrigel coating solution (see Recipes)
hiPSC culture medium (see Recipes)
EB mesoderm differentiation medium-1 (see Recipes)
EB mesoderm differentiation medium-2 (see Recipes)
Myelomonocytic expansion medium (see Recipes)
FACS buffer (see Recipes)
Osteoclast differentiation medium (see Recipes)
TRAP staining solution (see Recipes)
Reconstitution of hBMP4 (see Recipes)
Reconstitution of other cytokines (see Recipes)
Table 1. Sequences of qPCR primers to detect marker gene expression for mesoderm differentiation and osteoclastogenesis

Equipment
Pipettes
Water bath
AirClean 600 PCR workstation (AirClean Systems, model: AC648A )
Stereomicroscope (Carl Zeiss, model: Stemi 508 )
Cell culture incubator (Thermo Fisher Scientific, model: Heracell 240i, catalog number: 51026332)
Eppendorf refrigerated centrifuge (Eppendorf, model: 5810R)
CFX96 Touch Real-time PCR Detection System (Bio-Rad, catalog number: 1855196)
MACSQuant Analyzer 10 (Miltenyi Biotech, catalog number: 130-096-343)
Upright microscope (Carl Zeiss, model: Axio Imager.D2m)
Tabletop scanning electron microscope (Hitachi, model: TM1000)
Software
CFX manager software (Bio-Rad, 18450000)
FlowJoTM software (BD Bioscience, https://www.flowjo.com/)
GraphPad Prism (GraphPad, https://www.graphpad.com/)
Fiji ImageJ (NIH, image.nih.gov)
ZEN microscope software (Zeiss, https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Chen, I. (2020). Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) into Osteoclasts. Bio-protocol 10(24): e3854. DOI: 10.21769/BioProtoc.3854.
分类
干细胞 > 多能干细胞 > 细胞分化
细胞生物学 > 细胞分离和培养 > 细胞分化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link