- EN - English
- CN - 中文
Dual sgRNA-based Targeted Deletion of Large Genomic Regions and Isolation of Heritable Cas9-free Mutants in Arabidopsis
基于双sgRNA的拟南芥大基因组区域靶向清除及可遗传的非Cas9突变体的分离
发布: 2020年10月20日第10卷第20期 DOI: 10.21769/BioProtoc.3796 浏览次数: 6751
评审: Caiji GaoMatthew A. EscobarLifeng Liu
Abstract
CRISPR/Cas9 system directed by a gene-specific single guide RNA (sgRNA) is an effective tool for genome editing such as deletions of few bases in coding genes. However, targeted deletion of larger regions generate loss-of-function alleles that offer a straightforward starting point for functional dissections of genomic loci. We present an easy-to-use strategy including a fast cloning dual-sgRNA vector linked to efficient isolation of heritable Cas9-free genomic deletions to rapidly and cost-effectively generate a targeted heritable genome deletion. This step-by-step protocol includes gRNA design, cloning strategy and mutation detection for Arabidopsis and may be adapted for other plant species.
Keywords: CRISPR/Cas9 (CRISPR/Cas9)Background
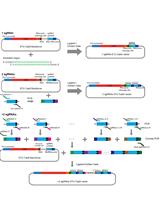
Dual sgRNA-directed gene knockout by CRISPR/Cas9 has been successfully used for genome editing in a variety of organisms (Wang et al., 2013; Chen et al., 2014; Char et al., 2017; Cai et al., 2018; Durr et al., 2018; Cui et al., 2019; Do et al., 2019; Liu et al., 2020). Targeted deletions of genomic DNA regions offer a valuable starting point for functional genomics studies (Hilton and Gersbach, 2015; Ford et al., 2019; Gowthaman et al., 2020). CRISPR/Cas9-based methods to delete genomic regions benefit from two gRNAs flanking the target DNA regions (Xiao et al., 2013; Canver et al., 2014; Kistler et al., 2015; Song et al., 2016). In plants, one key bottleneck to perform multiplex gene targeting from a single transformation event is to include multiple gBlocks into one binary vector. A gBlock is composed of a RNA Polymerase III (RNAPIII) promoter, a gene specific sgRNA protospacer, a sgRNA scaffold and a RNAPIII terminator. However, gBlock DNA sequences are usually long and repetitive, rendering design expensive for synthetic DNA and laborious by traditional assembly methods (Gao et al., 2016; Peterson et al., 2016; Zhang et al., 2016; Char et al., 2017; Durr et al., 2018; Pauwels et al., 2018; Schuster, 2018; Wu et al., 2018; Hui et al., 2019; Fonseca et al., 2020). For example, Durr et al. (2018) developed a dual-sgRNA vector by first modifying a pEN-Chimera entry vector to generate two gBlocks, then inserting two gRNAs into the modified entry vector by restriction enzymes, respectively, and finally cloning two gBlocks into the binary vectors. The multiple steps necessary are laborious and time-consuming. Although multiplex CRISPR/Cas9 platforms by introducing repetitive gBlocks for targeted genome editing were reported (Ordon et al., 2016; Schuster, 2018), several studies have noted that transformation of highly repetitive DNA sequences can trigger recombination and silencing of the RNA expression cassettes in a variety of species (Ma and Mitra, 2002; Lovett et al., 2004; Brake et al., 2008). To simplify targeted genomic regions and minimize potential recombination in Arabidopsis, we combined and modified existing cloning-based assembly steps. First, we amplified the middle border of our target specific two-gBlocks from a previously developed vector pHEE2E-TRI (Wang et al., 2015) in a single step. Second, we cloned the middle border into a known CRISPR/Cas9 binary vector pKIR1.1 (Maruyama et al., 2013; Tsutsui and Higashiyama, 2017), allowing two-gBlocks with different Poll III-dependent promoters to reduce the repetitiveness. This vector harbors an RPS5A-Cas9 cassette driving high constitutive expression of Cas9 protein at all developmental stages including egg cells, thus achieves highly efficient mutation in the T1 generation of Arabidopsis. In addition, the expression cassette OLE1-tagRFP in this system shows red fluorescence in seeds, allowing rapid screening for heritable Cas9-free Arabidopsis mutants in the seed of primary transformants. We combined the advantages of the two vectors by a single PCR and a single cloning step, thus providing a simple and reliable protocol to generate stable inherited deletion mutants. Our strategy promises to save cost and time to delete any chromosomal region in Arabidopsis, and can be likely adapted for genome editing of multiple genes simultaneously. It also has the potential to simplify genomic deletion in other plant species.
Materials and Reagents
- Consumables
- Sterile pipette tips (Axygen, catalog numbers: TF-300-R-S [10 µl], T-350-C-L-R-S [300 µl], TF-1000-R-S [1,000 µl])
- PCR microtubes (BioExpress, catalog number: T-3135-2 )
- 60 mm x 15 mm round Petri dishes (VWR, catalog number: 100488-404 )
- Competent cells
- Escherichia coli HST08 competent cell (homemade, protocol 1), store at -80 °C
- Agrobacterium tumefaciens GV3101 competent cells (homemade, protocol 2), store at -80 °C
- Vectors
- pHEE2E-TRI (Addgene, catalog number: 71288 ), store at -20 °C
- pKIR1.1 (Addgene, catalog number: 85758 ), store at -20 °C
pHEE2E-TRI sequence: https://www.addgene.org/71288/sequences/
pKIR1.1 sequence: https://www.addgene.org/85758/sequences/ - Oligonucleotides 10 µM
Dual-sgRNA1_F: 5′-CACCTGCATACATTGN20(protospacer 1)GTTTTAGAGCTAGAAATAGC-3′
Dual-sgRNA2_R: 5′-CACCTGCATACAAACN20(protospacer 2 reverse complement)CAATCTCTTAGTCGA CTCTAC-3′
Mlo 1938: 5′-TCCCAGGATTAGAATGATTAGG-3′
Primer_F: 5′-TTCTCTCTTCGCTCTCTCGTAG-3′
Primer_R: 5′-GGCCCAAATACTCTTTTCCAAGAC-3′
Cas9_F: 5′-CAGCCGACAAGAAGTACAGC-3′
Cas9_R: 5′-ATGGTGGGGTACTTCTCGTG-3′ - Enzymes and buffers
- AarI (Thermo Fisher Scientific, catalog number: ER1581 ), store at -20 °C
- T4 DNA Ligase (NEB, catalog number: M0202L ), store at -20 °C
- T4 Polynucleotide Kinase (NEB, catalog number: M0201L ), store at -20 °C
- T4 DNA Ligase Reaction Buffer (10x) (NEB, catalog number: B0202S ), store at -20 °C
- Alkaline Phosphatase, Calf Intestinal (CIP) (NEB, catalog number: M0290 ), store at -20 °C
- Phusion High-Fidelity DNA Polymerase Kits (New England Biolabs, catalog number: M0530S ), store at -20 °C
- HotMaster Taq DNA Polymerase (VWR, catalog number: QUNT2200330 ), store at -20 °C
- Wizard® SV Gel and PCR Clean-Up System (Promega, catalog number: A9282 ), store at room temperature
- DNA plasmid kit (VWR, catalog number: D6943-02 ), store at room temperature
- Reagents
- MES (Sigma-Aldrich, catalog number: 4432-31-9 ), store at room temperature
- KOH (Sigma-Aldrich, catalog number: 1310-58-3 ), store at room temperature
- Sucrose (Sigma-Aldrich, catalog number: 57-50-1 ), store at room temperature
- Plant agar (Sigma-Aldrich, catalog number: 9002-18-0 ), store at room temperature
- Bacto agar (BD Biosciences, catalog number: 214030 ), store at room temperature
- Bacto tryptone (BD Biosciences, catalog number: 211699 ), store at room temperature
- Bacto yeast extract (BD Biosciences, catalog number: 212730 ), store at room temperature
- Sodium chloride (Fisher Scientific, catalog number: 7647-14-5 ), store at room temperature
- Murashige & Skoog medium (Duchefa Biochemie, catalog number: M524 ), store at 4 °C
- Antibiotics
- Spectinomycin (VWR, catalog number: 101454-196 ), store at -20 °C
- Rifampicin (VWR, catalog number: 13292-46-1 ), store in a dry and well-ventilated place
- Gentamycin (VWR, catalog number: 97062-974 ), store at 4 °C
- Kanamycin (VWR, catalog number: 25389-94-09 ), store at 4 °C
- Media (see Recipes)
- LB liquid medium + 100 mg/L spectinomycin (store at 4 °C for one month)
- LB agar plates + 100 mg/L spectinomycin (store at 4 °C for one month)
- LB liquid medium + 100 mg/L spectinomycin + 20 mg/L rifampicin + 25 mg/L gentamycin + 25 mg/L kanamycin (store at 4 °C for one month)
- LB agar plates + 100 mg/L spectinomycin + 20 mg/L rifampicin + 25 mg/L gentamycin + 25 mg/L
- ½ MS-medium plates
Equipment
- Pipettes (Thermo Scientific, FinnpipetteTM F2, catalog numbers: 4642010 [0.2-2 µl], 4642030 [1-10 µl], 4642060 [2-20 µl], 4642080 [20-200 µl], 4642090 [100-1,000 µl])
- Incubator (Thermo Scientific, catalog number: 51028132 )
- Shakers (Eppendorf, model: New BrunswickTM Innova® 44 , catalog number: M1282-0000)
- Stereo Fluorescence Microscope (Leica, model: M205FA )
- Gel Doc EQ System (Bio-Rad, Universal Hood II, model: BGDII )
- Heating blocks (Eppendorf, catalog number: T1317-1EA )
- ThermoMixer® C (Eppendorf, catalog number: 5382000015 )
- PCR Thermo Cycler (Bio-Rad, model: T100 , catalog number: 1861096 )
- Tabletop centrifuge (Thermo Fisher Scientific, catalog number: 75008801 )
- NanoDrop (Thermo Scientific, model: NanoDropTM 2000C , catalog number: ND2000)
- Agarose gel electrophoresis equipment (Bio-Rad, catalog number: 1704489EDU )
- Plant growth chamber (photoperiod: 16 h light/ 8 h dark, temperature: 22 °C in the day/20 °C in the darkness, humidity: 65%, light intensity: 100 μE m−2 s−1)
Note: No equipment from specific manufacturers is required. Any equivalent device can be used.
Software
- SnapGene® (SnapGene, https://www.snapgene.com/)
- Image Lab Software (Bio-Rad, https://www.bio-rad.com/)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Jin, Y. and Marquardt, S. (2020). Dual sgRNA-based Targeted Deletion of Large Genomic Regions and Isolation of Heritable Cas9-free Mutants in Arabidopsis. Bio-protocol 10(20): e3796. DOI: 10.21769/BioProtoc.3796.
分类
植物科学 > 植物分子生物学 > DNA > 诱/突变
植物科学 > 植物分子生物学 > 遗传分析
分子生物学 > DNA > 染色体工程
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link