- EN - English
- CN - 中文
Nonenzymatic RNA-templated Synthesis of N3′→P5′ Phosphoramidate DNA
N3'→P5'磷酰胺DNA的非酶RNA模板合成
(*contributed equally to this work) 发布: 2020年09月05日第10卷第17期 DOI: 10.21769/BioProtoc.3734 浏览次数: 4399
评审: Imre GáspárLouise Jane WalportKuo-Ching "KC" Mei
Abstract
The RNA world hypothesis describes a scenario where early life forms relied on RNA to govern both inheritance and catalyze useful chemical reactions. Prior to the emergence of enzymes capable of replicating the RNA genome, a nonenzymatic replication process would have been necessary to initiate Darwinian Evolution. However, the one-pot nonenzymatic RNA chemical copying of templates with mixed-sequences is insufficient to generate strand products long enough to encode useful function. The use of alternate (RNA-like) genetic polymers may overcome hurdles associated with RNA copying, and further our understanding of nonenzymatic copying chemistry. This protocol describes the nonenzymatic copying of RNA templates into N3′→P5′ phosphoramidate DNA (3′-NP-DNA). We describe, in detail, the synthesis of 3′-amino-2′,3′-dideoxyribonucleotide monomers activated with 2-aminoimidazole (3′-NH2-2AIpddN), and their use in template-directed polymerization.
Keywords: Nonenzymatic template-directed synthesis (非酶模板定向合成)Background
Primitive life forms could have consisted of a genome capable of replication and function, encapsulated within a spatially-defined compartment (Szostak et al., 2001; Szostak, 2012; Blain and Szostak, 2014). Prior to the emergence of enzymes capable of replicating the RNA genome, a nonenzymatic replication process would have been necessary to initiate Darwinian Evolution (Szostak, 2012 and 2017). RNA is a logical candidate for the primordial genetic polymer given its ability to catalyze critical chemical reactions such as protein synthesis, and to act as a genetic information carrier in modern biological contexts.
Despite major breakthroughs toward demonstrating nonenzymatic RNA replication over the past 5 decades, there remain major hurdles to overcome before achieving this seemingly overarching problem in origins of life research (Szostak, 2012 and 2017). Pioneering work by Leslie Orgel showed that high-energy nucleoside 5'-phosphoroimidazolides are capable of spontaneously polymerizing in a template-directed fashion (Szostak, 2017). Our group and others have used nonenzymatic primer extension by activated mononucleotides as a model for the study of prebiotic genome copying (Figure 1A). Recently, we showed that short activated oligoribonucleotides that can bind to an RNA template segment downstream of an activated ribonucleotide can catalyze the reaction on the monomer with the RNA primer (Prywes et al., 2016). In addition, by screening a small library of leaving groups, Li et al. discovered that 2-aminoimidazole (2AI) is a superior nucleotide activating group (Figure 1) (Li et al., 2017). These two improvements allowed copying of mixed-sequence RNA templates in one-pot for the first time. Nevertheless, the template-directed chemical copying into complementary RNA sequences remains limited to only seven nucleotides in solution (Li et al., 2017). As a result, we hypothesize that other factors can further optimize primer extension reaction, and that it may be possible to discover prebiotically plausible pathways to copy RNAs of sufficient length to encode useful biological function (typically 30-50 nucleotides in length). Toward this end, our group and others have explored alternative genetic materials, which may provide insights into how longer RNA sequences could be propagated nonenzymatically (Rojas Stütz and Richert, 2001; Zhang et al., 2013a and 2013b; Hänle and Richert, 2018). We chose N3′→P5′ phosphoramidate DNA (3′-NP-DNA, Figure 1B) as a potential proxy for RNA for a number of reasons. First, the 3′-NP-DNA biopolymer has a strong structural resemblance to the A-form RNA duplex (double stranded RNA) (Tereshko et al., 1998). Moreover, it hybridizes with RNA and DNA with higher thermal stability than the homo-duplexes (Gryaznov et al., 1995; Gryaznov, 1997), and 3′-amino-2′,3′-dideoxyribonucleotides (Figure 1B, the monomeric units that polymerize into 3′-NP-DNA) adopt a C3′-endo conformation in solution similar to ribonucleotides (Ding et al., 1998; Zhang et al., 2012). The latter feature is known to speed up nonenzymatic primer extension reactions (Zhang et al., 2012).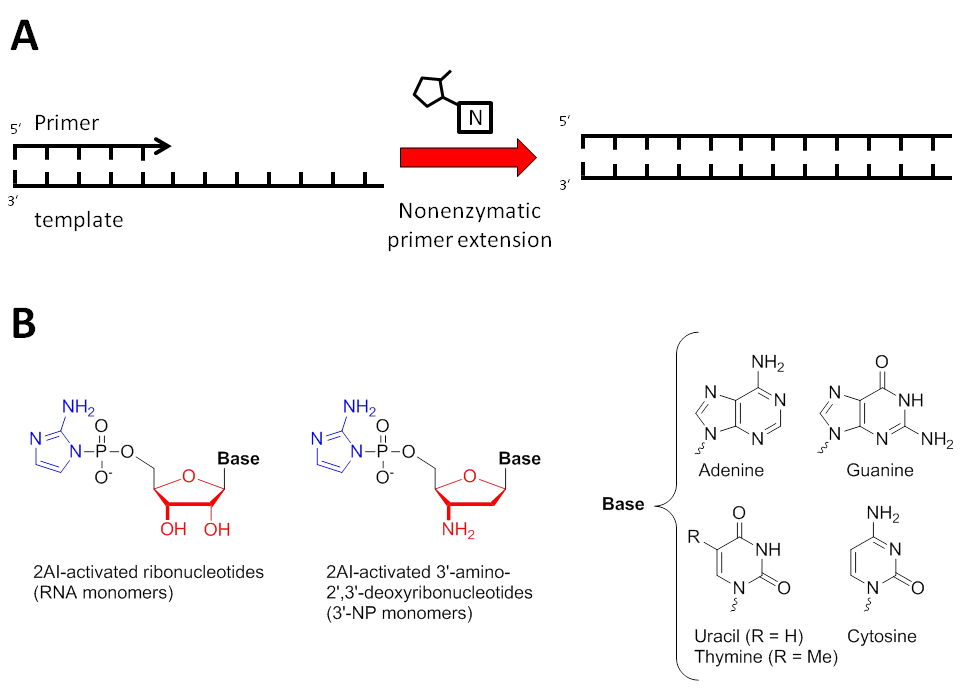
Figure 1. Nucleic acid primer extension model. A. Schematic representation of primer extension experiments. Activated mononucleotides are indicated above the arrow and participate in nonenzymatic primer extension. B. Chemical structure of ribonucleotides and 3′-amino-2′,3′-dideoxyribonucleotides (3′-NP) activated with 2-aminoimidazole (2AI) (3′-NH2-2AIpddN, where N signifies the identity of the nucleobase). The 2AI leaving groups are highlighted in blue, and the furanose moieties are highlighted in red to illustrate the chemical differences between ribose and 3′-amino-2′,3′-dideoxyribose. Note that conventionally, thymine is used for the 3′-NP system (as opposed to uracil, which is found in RNA).
There are few detailed protocols for performing nonenzymatic primer extension, including the preparation of activated 5′-phosphorimidazolides (e.g., Figure 1B). Recently, our group contributed a manuscript, in video format, concerning the preparation and use of fatty acid-containing vesicles, with a section detailing the procedures for nonenzymatic RNA copying within vesicles (Jin et al., 2018). The current protocol describes the procedures for primer extension experiments in much more detail, including the synthesis of the activated mononucleotides, synthesis of oligomers, and primer extension reactions. This protocol may be used in the origins of life field and for research concerning the bottom-up approach to building an artificial cell.
Materials and Reagents
- Disposable Norm-ject syringe (Thermo Fisher Scientific, 1 ml, 5 ml, 10 ml, 20 ml)
- Disposable needle 18G (Thermo Fisher Scientific, catalog number: 14-840-96 )
- Syringe filter (Thermo Fisher Scientific, catalog number: SLGVM33RS )
- pH-indicator strips pH 0-14 Universal indicator (Millipore Sigma, catalog number: 1095350001 )
- 1.5 ml Microcentrifuge tubes (Bag of 500) (conical bottom, Millipore Sigma, catalog number: 1000-0785 )
- 2.0 ml FisherbrandTM Free-Standing Microcentrifuge Tubes with Screw Caps (Thermo Scientific, catalog number: 02-682-558 )
- Filter paper (Whatman, to fit Buchner funnel)
- 3'-Amino-2',3'-dideoxyguanosine, 99% (Alfa Aesar, catalog number: J65326 )
- 3'-Amino-2',3'-dideoxythymidine, 99% (Alfa Aesar, catalog number: J64342 )
- 3'-Amino-2',3'-dideoxyadenosine, 98% (Alfa Aesar, catalog number: J65090 )
- 3'-Amino-2',3'-dideoxycytidine (Biosynth Carbosynth, catalog number: NA05183 )
- 3'-(TFA)amino-2',3'-dideoxycytidine 5'-CED phosphoramidite (ChemGenes, catalog number: ANP-1242 )
- Reverse RNA phosphoramidites:
2'-TBDMS-3'-DMT-rA (N-bz) (ChemGenes, catalog number: AN-3401 )
2'-TBDMS-3'-DMT-rC (N-acetyl) (ChemGenes, catalog number: AN-3405 )
2'-TBDMS-3'-DMT-rG (N-iPr PAC) (ChemGenes, catalog number: AN-3406 )
2'-TBDMS-3'-DMT-rU (ChemGenes, catalog number: AN-3404 ) - 3'-(6-FAM) CPG (Glen Research, catalog number: 20-2961-41E for ExpediteTM 8900 Nucleic Acid Synthesis System)
- 9-Fluorenylmethyl N-succinimidyl carbonate, Fmoc-OSu (Combi-Blocks, catalog number: ST-5099 )
- Pyridine (Py) (Millipore Sigma, catalog number: 270970-100ML )
- Dichloromethane (DCM) (Millipore Sigma, catalog number: 320269-4L )
- Diethylether (Et2O) (Millipore Sigma, catalog number: 296082-2.5L )
- Sodium perchlorate (NaClO4) (Millipore Sigma, catalogue number: 410241-500G)
- Acetone (Millipore Sigma, catalog number: 179124-4L )
- Molecular Sieves 3A (Thermo Fisher Scientific, catalog number: AC197255000 )
- N,N-Dimethylformamide (DMF) (Millipore Sigma, catalog number: 227056-100ML )
- Dimethylsulfoxide, anhydrous (DMSO) (Millipore Sigma, catalog number: 276855-100ML )
- Dimethyl sulfoxide-d6 (d6-DMSO) (Millipore Sigma, catalog number: 156914-25G )
- Trimethylphosphate (TMP) (Thermo Fisher Scientific, catalog number: AC157970500 )
- N,N-Diisopropylethylamine (DIPEA) (Millipore Sigma, catalog number: 387649-100ML )
- Triethylamine (TEA) (Millipore Sigma, catalog number: 8083521000 )
- Acetonitrile, anhydrous (MeCN) (Glen Research, catalog number: 40-4050-50 )
- Acetonitrile, DNA synthesis grade (MeCN) (Thermo Fisher Scientific, catalog number: A21 )
- Phosphorus (V) oxychloride, 99% (POCl3) (Thermo Fisher Scientific, catalog number: AA1052530 )
- Dry ice (solid carbon dioxide)
- 2-Amino-1H-imidazole, HCl (2AI·HCl) (Combi-Blocks, catalog number: SS-6610 )
- Triphenylphosphine (TPP) (Millipore Sigma, catalog number: 8082700250 )
- 2,2'-Dipyridyldisulfide (DPDS) (Combi-Blocks, catalog number: QA-4326 )
- Piperidine (Millipore Sigma, catalog number: 411027-100ML )
- Sodium hydroxide (NaOH) (50% solution in water) (Millipore Sigma, catalog number: 415413-100ML )
- Triethylamine trihydrofluoride (NEt3·3HF) (Millipore Sigma, catalog number: 344648-25G )
- Sodium acetate (NaOAc) (Millipore Sigma, catalog number: S2889-250G )
- EDTA solution (500 mM, pH 8, Thermo Fisher Scientific, catalog number: AM9260G )
- Xylene cyanol FF (Millipore Sigma, catalog number: X4126-10G )
- Bromophenol blue (Millipore Sigma, catalog number: B0126-25G )
- Tris-Borate-EDTA buffer (TBE) (Millipore Sigma, catalog number: T4415-1L )
- Urea (Millipore Sigma, catalog number: U5378 )
- Ethanol 200 proof (EtOH) (Decon Labs, catalog number: 2716 )
- Methanol (MeOH) (Thermo Fisher Scientific, catalog number: A412 )
- Chloroform (CHCl3) (Millipore Sigma, catalog number: 472476-2.5L )
- Sodium chloride (NaCl) (Millipore Sigma, catalog number: S7653-250G )
- Magnesium chloride (MgCl2) (Thermo Fisher Scientific, catalog number: AM9530G )
- NH4OH solution (28% NH3 in water, Millipore Sigma, catalog number: 338818-100ML )
- MeNH2 solution (40% wt. in water, Millipore Sigma, catalog number: 426466-100ML )
- SequaGel (UreaGel Concentrate) (National Diagnostics, catalog number: EC-830 ) to make 19:1 (w/w) acrylamide:bis-acrylamide
- SequaGel (UreaGel Diluent) (National Diagnostics, catalog number: EC-840 ) to make 19:1 (w/w) acrylamide:bis-acrylamide
- N,N,N′,N′-Tetramethylethylenediamine (TEMED) (Millipore Sigma, catalog number: T9281-50ML )
- 1 M Na+-HEPES pH 8 (Thermo Fisher Scientific, catalog number: AAJ63578AK )
- Ammonium persulfate (APS) (Millipore Sigma, catalog number: A3678-100G )
- Sep-Pak C18 Plus Short Cartridge, 360 mg (Waters, catalog number: WAT020515 )
- RNA template (Integrated DNA technologies, HPLC purified)
- RNA primer containing 3'-NH2 terminal (synthesized in house, see Procedure D)
Equipment
- Analytical balance (e.g., SartoriusTM SecuraTM Analytical Weighing Balances, catalog number: 14-557-401 )
- Round bottom flask (25 ml, 50 ml, 100 ml, 500 ml, Chemglass Life Sciences)
- Septum stoppers for round bottom flasks (Chemglass Life Sciences)
- Vacuum filtration setup (Buchner funnel, Erlenmeyer filter flask, filter adaptors (neoprene), Chemglass Life Sciences)
- Water bath (e.g., large Erlenmeyer flask, Chemglass Life Science)
- Single-channel pipettor (1,000 μl, 200 μl, 20 μl, 2 μl, Thermo Fisher Scientific)
- Pipet Controller (Thermo Fisher Scientific, e.g., BrandTechTM accu-jetTM pro, catalog number: 03-840-311 )
- Stir bar (e.g., 19 mm (3/4”) x 19 mm (3/4”) x 9.5 mm (3/8”), VWR, catalog number: 58947-822 )
- Rotary evaporator (Heidolph Laborota 4001 efficient)
- Low-resolution mass spectrometry (Bruker Esquire 6000)
- Combi Flash Rf-200 automated flash chromatography system (Teledyne Isco, part number: 68523006)
- RediSep Rf columns (Teledyne Isco, catalog numbers: 69-2203-340 (40 g) and 69-2203-380 (80 g))
- RediSep Rf Gold C18 Column (Teledyne Isco, catalog number: 69-2203-336 )
- Benchtop centrifuge (Eppendorf, model: 5910R )
- Razor blade (FisherbrandTM, catalog number: 12-640 )
- Speedvac concentrator (Thermo Scientific Savant SPD111V)
- Digital block heater (VWR, catalog number: 12621-088 )
- NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, catalog number: DN-2000C )
- Nucleic acid automated synthesizer Expedite 8909
- Thermal cycler (Bio-Rad, T100TM Thermal Cycler)
- Electrophoresis power supply (Bio-Rad, catalog number: 164-5056 )
- 0.75 mm thick, 1.5 mm thick spacers (Millipore Sigma, catalog numbers: EP1418-2EA , EP1420-2EA , respectively)
- 30-well VS20 comb (Millipore Sigma, catalog number: EP1428 )
- Preparatory comb (Apogee Electrophoresis, catalog number: 21076021 )
- Vertical electrophoresis system glass plates (Apogee Electrophoresis, catalog number 11074010)
- Electrophoresis glass plates (15 x 17 cm vertical gel plates, Apogee Electrophoresis)
- Amersham Typhoon RGB Biomolecular Imager (GE Healthcare Life Sciences, catalog number: 29187193 )
- Nuclear Magnetic Resonance (NMR, 400MHz Variant)
Software
- ChemDraw (PerkinElmer)
- ImageQuantTM (GE Healthcare Life Sciences)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
O'Flaherty, D. K., Zhou, L. and Szostak, J. W. (2020). Nonenzymatic RNA-templated Synthesis of N3′→P5′ Phosphoramidate DNA. Bio-protocol 10(17): e3734. DOI: 10.21769/BioProtoc.3734.
分类
分子生物学 > RNA > 返转录
分子生物学 > DNA > DNA 合成
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link