- EN - English
- CN - 中文
A SsrA/NIa-based Strategy for Post-Translational Regulation of Protein Levels in Gram-negative Bacteria
基于SsrA/NIa的革兰阴性菌蛋白质水平翻译后调控策略
发布: 2020年07月20日第10卷第14期 DOI: 10.21769/BioProtoc.3688 浏览次数: 4380
评审: Alba BlesaLi ZhangThibaud T. Renault
Abstract
Strategies to control the levels of key enzymes of bacterial metabolism are commonly based on the manipulation of gene of interest within the target pathway. The development of new protocols towards the manipulation of biochemical processes is still a major challenge in the field of metabolic engineering. On this background, the FENIX (functional engineering of SsrA/NIa-based flux control) system allows for the post-translational regulation of protein levels, providing both independent control of the steady-state protein amounts and inducible accumulation of target proteins. This strategy enables an extra layer of control over metabolic fluxes in bacterial cell factories (see Graphical abstract below). The protocol detailed here describes the steps needed to design FENIX-tagged proteins and to adapt the system to virtually any pathway for fine-tuning of metabolic fluxes.
Graphical abstract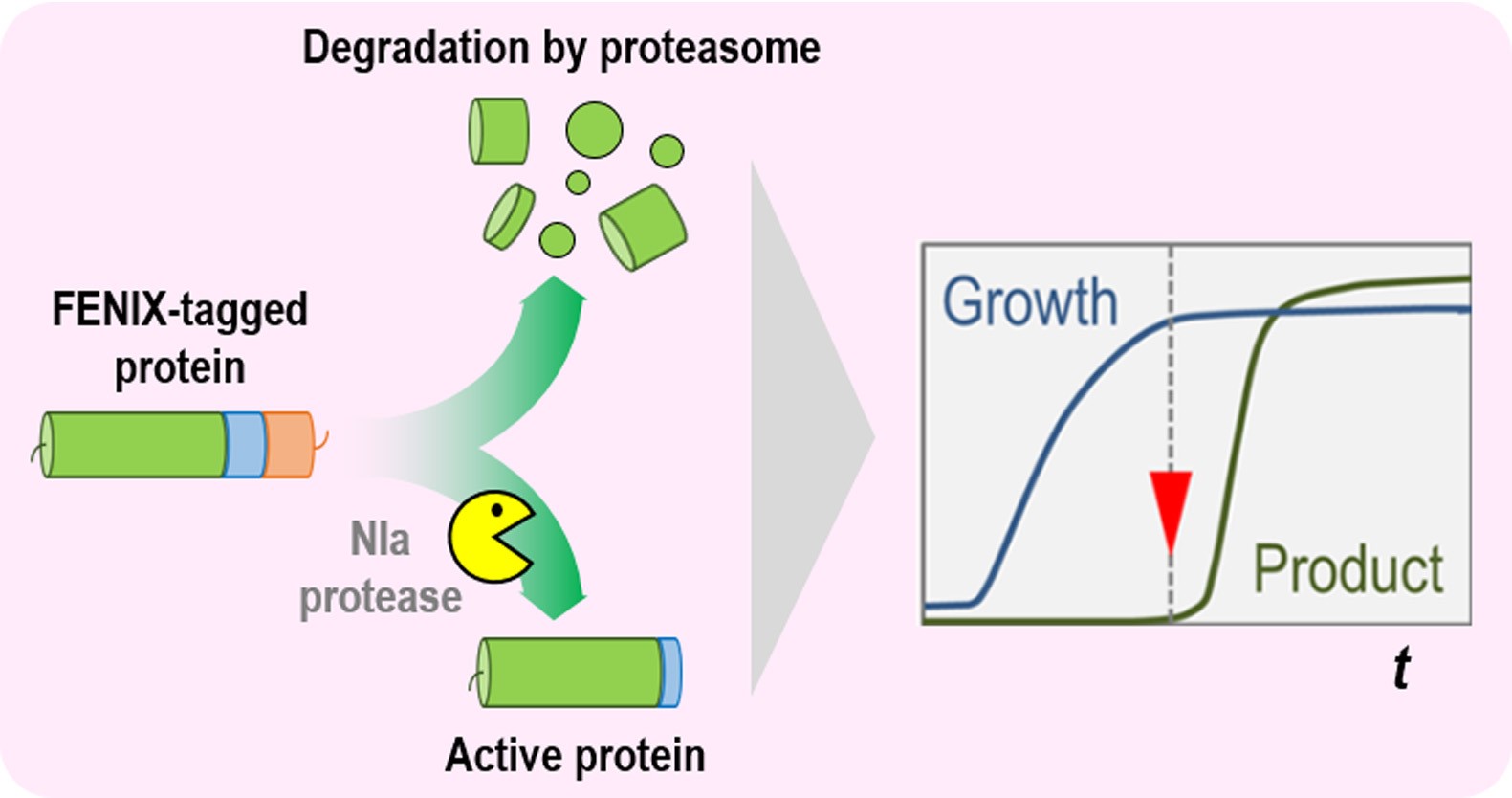
Background
Controlling protein production has become a challenging problem that has been mostly tackled by manipulating the regulation of its production at different levels, such as the DNA level or the amount of protein(s) produced (Wu et al., 2016; Avcilar-Kucukgoze et al., 2017). At the DNA level, both transcription and translation have been widely studied in several microorganisms, enabling the design and development of a large number of synthetic circuits to improve bioproduction processes (Guzmán et al., 1995; Lutz and Bujard, 1997). In contrast, the protein level has been much less studied, with protocols mainly based on RNA interference, riboregulators and specific transcriptional regulators (Isaacs et al., 2004; Lou et al., 2012).
In order to implement a new approach towards the regulation of protein levels, we recently proposed a synthetic biology tool called FENIX (functional engineering of SsrA/NIa-based flux control) (Durante-Rodríguez et al., 2018). FENIX is based on the activity of the protease NIa, capable of recognizing and cleaving a specific sequence of 13 amino acids (the nia target, GESNVVVHQADER) (Verchot et al., 1991; Stevens, 2000; Calles et al., 2019), and the action of the proteasome, which recognizes a sequence of 11 amino acids located in the C-terminal region of a protein (the ssrA target, AANDENYALAA) (Doma and Parker, 2007; Shoemaker et al., 2010). We have designed a synthetic NIa/SsrA tag (GESNVVVHQADER•AANDENYALAA) that can be easily fused to the C-terminal module of any protein of interest via a single cloning step in a standardized vector. The FENIX system relies on the constitutive degradation of the target protein by the proteasome, followed by conditionally restoring protein accumulation by the cleavage of the proteasome tag (ssrA) through the action of the protease NIa. The whole circuit is triggered by the addition of 3-methylbenzoate, which induces the XylS/Pm expression system, thus controlling the individual components of the FENIX system. This novel tool can be also used in Pseudomonas putida for tight control of protein accumulation (Volke et al., 2020). The protocol described below discusses the overall strategy along with specific details on its implementation.
Materials and Reagents
- Kits
- High-Pure plasmid isolation kit (Roche, catalog number: 11754777001 )
- Gene-Clean Turbo kit (Q-BIOgene, catalog number: 1102-400 )
- Enzymes
- Pfu DNA polymerase (Promega, catalog number: M7741 )
- T4 DNA ligase (New England Biolabs, catalog number: M0202L )
- Restriction enzyme NheI (New England Biolabs, catalog number: R3131S )
- Restriction enzyme BsrGI (New England Biolabs, catalog number: R3575S )
- Restriction enzyme SpeI (New England Biolabs, catalog number: R3133L )
- Chemicals and other consumables
- 0.22 μm filter membranes (Merck, catalog number: GSWP02500 )
- Multiwell microtiter plates, 96-well, Flat Bottom (Falcon, catalog number: 353072 )
- 50 ml plastic tubes (Falcon)
- 2-ml microcentrifuge tubes
- 1.5-ml microcentrifuge tubes
- Aluminum foil
- Petri dishes
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378-25G )
- Kanamycin sulfate (Roche, catalog number: 10106801001 )
- Absolute ethanol (Sigma-Aldrich, catalog number: 1009832500 )
- 85% (v/v) glycerol (Sigma-Aldrich, catalog number: 1040942500 )
- NaCl (Merck, catalog number: 1.06404.1000 )
- NaOH (Merck, catalog number: 1.06498.0500 )
- CaCl2 (Merck, catalog number: 1.02382.1000 )
- RbCl (Sigma-Aldrich, catalog number: R2252-100G )
- MnCl2 (Merck, catalog number: 1.05927.0100 )
- Potassium acetate (Merck, catalog number: 1.04820.1000 )
- Acetic acid (glacial) (Merck, catalog number: 1.00063.1000 )
- MOPS (Sigma-Aldrich, catalog number: M1254-250G )
- 3-methylbenzoate (Sigma-Aldrich, catalog number: 89890 )
- Agar (Conda, catalog number: 1806 )
- Agarose D1 (Conda, catalog number: 8019.22 )
- BactoTM tryptone (BD, catalog number: 211705 )
- BactoTM yeast (BD, catalog number: 212750 )
- Competent E. coli DH10B cells (see Recipes)
- Solution for DH10B competent cells TBFI (see Recipes)
- Solution for DH10B competent cells TBFII (see Recipes)
- 30 mg/ml chloramphenicol and 50 mg/ml kanamycin (see Recipes)
- LB broth (see Recipes)
- LB agar with 30 μg/ml chloramphenicol and/or 50 μg/ml kanamycin (see Recipes)
- 0.5 M 3-methylbenzoate (see Recipes)
- 50% (v/v) and 70% (v/v) ethanol (see Recipes)
Equipment
- ABI Prism 377 automated DNA sequencer (Applied Biosystems Inc.)
- Electroporator (Gene Pulser, Bio-Rad)
- Orbital shaker
- Flasks (50 ml, 1 L)
- Pipettes (Gilson, models: PIPETMAN P2, P10, P20, P100, P200, P1000, P5000)
- Centrifuge (Tomy, model: MX-301 )
- Thermal Cycler (Applied Biosystems, model: 2720Thermal Cycler )
- Temperature chamber (Taitec, model: Thermo minder SM-10R )
- Water bath shaker (Taitec, model: Personal-11 )
- Plate reader (Corona, model: MTP-880Lab )
- Autoclave (Tomy Seiko, model: LSX-500 )
Software
- Microsoft Excel (Microsoft)
- BioEdit Sequence Alignment Editor
- ApE-A plasmid Editor (https://jorgensen.biology.utah.edu/wayned/ape)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Durante-Rodríguez, G., Calles, B., Lorenzo, V. D. and Nikel, P. I. (2020). A SsrA/NIa-based Strategy for Post-Translational Regulation of Protein Levels in Gram-negative Bacteria. Bio-protocol 10(14): e3688. DOI: 10.21769/BioProtoc.3688.
分类
微生物学 > 微生物新陈代谢 > 其它化合物
微生物学 > 微生物生物化学 > 蛋白质
分子生物学 > 蛋白质 > 靶向降解
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link














