- EN - English
- CN - 中文
Genomic Edition of Ashbya gossypii Using One-vector CRISPR/Cas9
利用一个CRISPR/Cas9载体对棉囊阿舒氏酵母进行基因组编辑
(*contributed equally to this work) 发布: 2020年06月20日第10卷第12期 DOI: 10.21769/BioProtoc.3660 浏览次数: 4796
评审: Samantha E. R. DundonMoon ChatterjeeJulie Weidner
Abstract
The CRISPR/Cas9 system is a novel genetic tool which allows the precise manipulation of virtually any genomic sequence. In this protocol, we use a specific CRISPR/Cas9 system for the manipulation of Ashbya gossypii. The filamentous fungus A. gossypii is currently used for the industrial production of riboflavin (vitamina B2). In addition, A. gossypii produces other high-value compounds such as folic acid, nucleosides and biolipids. A large molecular toolbox is available for the genomic manipulation of this fungus including gene targeting methods, rapid assembly of heterologous expression modules and, recently, a one-vector CRISPR/Cas9 editing system adapted for A. gossypii that allows marker-free engineering strategies to be implemented. The CRISPR/Cas9 system comprises an RNA guided DNA endonuclease (Cas9) and a guide RNA (gRNA), which is complementary to the genomic target region. The Cas9 nuclease requires a 5′-NGG-3′ trinucleotide, called protospacer adjacent motif (PAM), to generate a double-strand break (DSB) in the genomic target, which can be repaired with a synthetic mutagenic donor DNA (dDNA) by homologous recombination (HR), thus introducing a specific designed mutation. The CRISPR/Cas9 system adapted for A. gossypii largely facilitates the genomic edition of this industrial fungus.
Keywords: CRISPR/Cas9 (CRISPR/Cas9)Background
The CRISPR/Cas9 system has emerged as a powerful tool for genome engineering (Horvath et al., 2010). A CRISPR/Cas9 system comprises an RNA guided Cas9 endonuclease that produces DSBs at specific genomic loci. The Cas9 associated RNA is formed by a CRISPR targeting RNA (crRNA) and a transactivating crRNA (tracrRNA), which are required to form the catalytic active Cas9 (Jinek et al., 2012). Both the crRNA and tracrRNA can be combined in a synthetic guide RNA (sgRNA) (Jinek et al., 2012). The gRNA determines the target loci and the Cas9 nuclease requires the presence of a PAM sequence in the genomic target to generate a DSB (Pattanayak et al., 2013). There are two mechanisms for DSB repair: homologous recombination (HR) and non-homologous end joining (NHEJ). The presence of a synthetic dDNA provides the ability to repair a targeted DSB by HR (Ran et al., 2013).
The CRISPR/Cas9 systems need to be adapted for each organism in order to achieve an efficient expression of both the gRNA and Cas9. For the A. gossypii system, a one-vector strategy was followed to contain all the required modules for CRISPR/Cas9 functionality (Figure 1): Cas9 expression, sgRNA expression and dDNA modules. The CAS9 expression module that was previously reported in Saccharomyces cerevisiae was used, where the human codon-optimized Streptococcus pyogenes CAS9 gene is under the control of the yeast TEF1 promoter and CYC1 terminator sequences (Dicarlo et al., 2013). In addition, an sgRNA expression module was designed under the control of the promoter and terminator sequences from the A. gossypii SNR52 gene, which is transcribed by RNA polymerase III. The sgRNA contained two sequences: a 20 bp sequence that targets a selected genomic locus and a 79 bp sequence for Cas9 binding (Figure 1). Also, the A. gossypii system contained the dDNA for DSB repair by HR (Figure 1). The assembly of the CRISPR/Cas9 vector with a specific synthetic gRNA-dDNA is carried out using a directional cloning strategy (Jiménez et al., 2019). Therefore, for each specific genomic edition, it is only necessary to design a specific sgRNA-dDNA both for gene targeting and DNA repair by HR. The CRISPR/Cas9 vector also contained a loxP-KanMX-loxP marker (G418R) for plasmid selection in A. gossypii (Jiménez et al., 2019).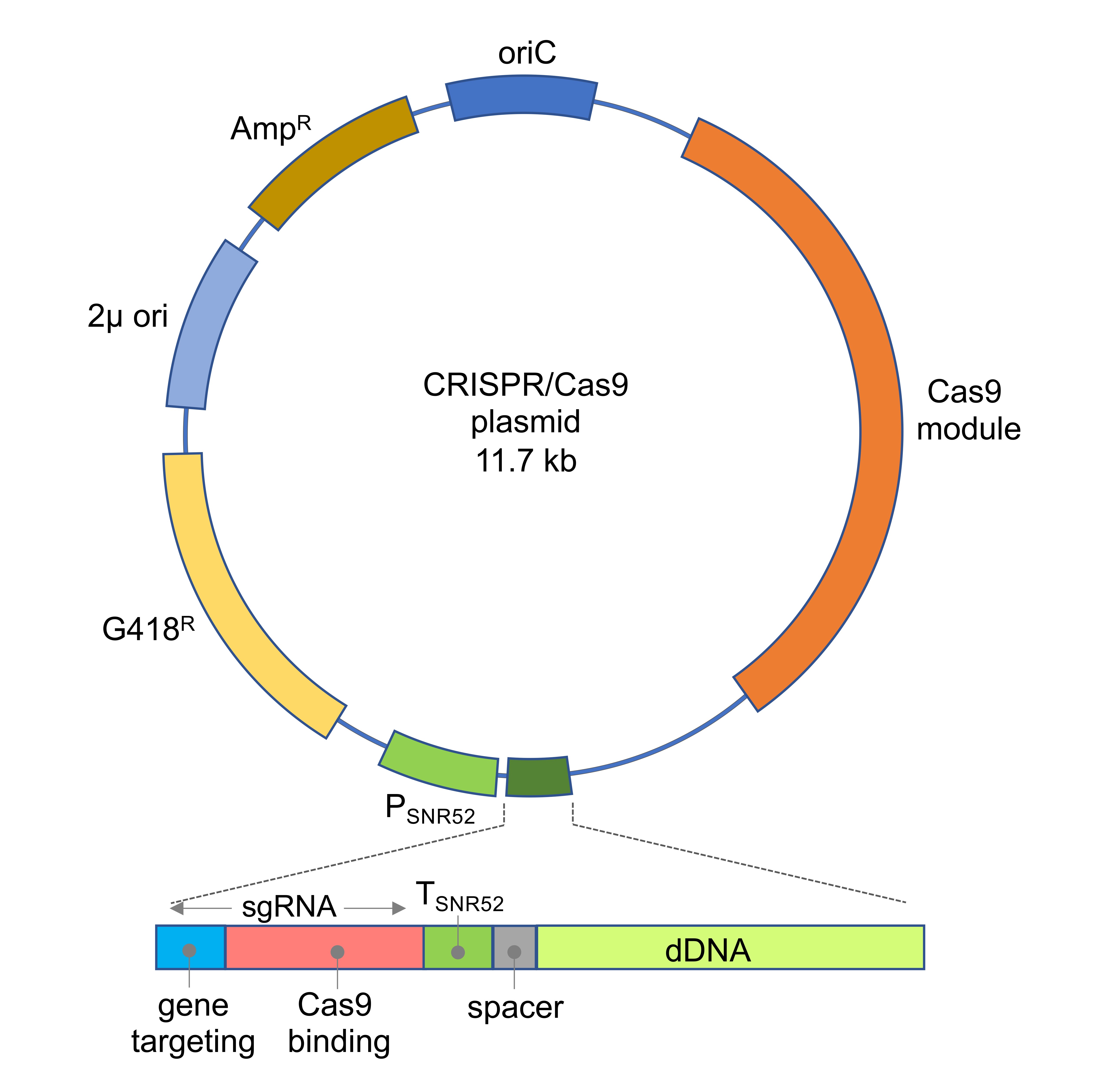
Figure 1. Map of the CRISPR/Cas9 plasmid. The sgRNA-dDNA module is depicted: the sgRNA comprises both sequences for gene targeting (blue) and Cas9 binding (red). The expression of the sgRNA is driven by the promoter and terminator sequences of SNR52. A short spacer sequence (grey) links the sgRNA module with the dDNA module (light green).
Most of the CRISPR/Cas9 systems require two transformation events: one for the introduction of the Cas9 and gRNA modules and a second one for the introduction of dDNA (DiCarlo et al., 2013). This strategy hinders the efficiency of the system in a multinucleated syncytium such as the A. gossypii mycelia. Hence, the use of a one-vector strategy in the A. gossypii CRISPR/Cas9 system largely increases its efficiency (Jiménez et al., 2019). Indeed, an average editing efficiency of 60% is achieved with the A. gossypii CRISPR/Cas9 system (Jiménez et al., 2019).
The CRISPR/Cas9 system for A. gossypii represents a novel methodology for marker-less gene deletions, insertions and nucleotide substitutions. Also, multiplexing CRISPR/Cas9 engineering can be achieved for the simultaneous edition of different targets and metabolic pathways, as previously demonstrated for S. cerevisiae (Bao et al., 2015; Jakociunas et al., 2015). In summary, the CRISPR/Cas9 system for A. gossypii will help with the rapid expansion of precise and efficient genomic editing strategies for this microorganism with industrial interest.
Materials and Reagents
Note: Materials and reagents can be purchased from different suppliers. Here, we show one list of all reagents and materials that we used to develop the protocol.
Materials
- Pipette tips (e.g., VWR, catalog numbers: 613-0340 , 613-0360 , 613-1068 )
- PCR microtubes (e.g., Sarstedt, catalog number: 72.737 )
- 1.5 ml microtube (Deltalab, catalog number: 200400P )
- 50 ml centrifuge tube (Corning, catalog number: 352070 )
- Graduated pipettes 10 ml (Brand, catalog number: 27011 )
- Petri dishes (Thermo Scientific, catalog number: 11309283 )
- Microscope slides (e.g., VWR, catalog number: 630-2099 )
- Cover slides (e.g., VWR, catalog number: 631-0125 )
- 0.4 cm electroporation cuvettes (Bio-Rad, catalog number: 1652088 )
- Syringe 50 ml (Omnifix, catalog number: 4616502F )
- Sterile syringe filter w/0.45 µm cellulose acetate membrane (VWR, catalog number: 28145-481 )
- Filter paper
Reagents
- PCR primers (Invitrogen), desalted, standard 25-50 nmol scale
- Escherichia coli DH5α (Thermo Fisher Scientific, catalog number: 18265017 )
- D(+)-Glucose monohydrate (Acros Organics, catalog number: 450740050 )
- D(+)-Sucrose (PanReac AppliChem, catalog number: 131621.1211 )
- Peptone (Condalab, catalog number: 1616.05 )
- Tryptone (Condalab, catalog number: 1612.05 )
- Yeast extract (Condalab, catalog number: 1702.05 )
- Malt extract (VWR, catalog number: J873-500G )
- Myo-inositol 99% (Sigma-Aldrich, CAS number: 87-89-8 )
- Soybean oil (Santa Cruz Biotechnology, catalog number: 8001-22-7 )
- Corn steep liquor (Sigma-Aldrich, CAS number: 66071-94-1 )
- European bacteriological agar (Condalab, catalog number: 1800.05 )
- Tris Base (Fisher BioReagents, catalog number: BP152-500 )
- Boric acid (PanReac AppliChem, catalog number: 131015 )
- EDTA (Ethylenediaminetetraacetic acid disodium salt 2-hydrate) (PanReac AppliChem, catalog number: 131669 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, CAS number: 7791-18-6 )
- Sodium chloride (NaCl) (PanReac AppliChem, catalog number: 131659.1214 )
- Potassium phosphate monobasic (KH2PO4) (Fisher BioReagents, CAS number: 7778-77-0 )
- Potassium phosphate dibasic (K2HPO4) (Fisher BioReagents, CAS number: 7758-11-4)
- Sodium carbonate anhydrous (Na2CO3) (PanReac AppliChem, catalog number: 131648.1211 )
- Sodium hydrogen carbonate (NaHCO3) (PanReac AppliChem, catalog number: 141638.121 0)
- Triton X-100 (Acros Organics, catalog number: 215680010 )
- DTT (dithiothreitol) (Thermo Scientific, catalog number: R0861 )
- PVP 40 (Polyvinylpyrrolidone) (Sigma-Aldrich, CAS number: 9003-39-8 )
- Tween 20 (PanReac AppliChem, catalog number: 142312.1611 )
- Bovine Serum Albumin (Sigma-Aldrich, CAS number: 9048-46-8 )
- G-418 Disulphate (Sigma-Aldrich, CAS number: 108321-42-2 )
- Kanamycin sulfate (Sigma-Aldrich, CAS number: 25389-94-0 )
- Ampicillin sodium salt (Sigma-Aldrich, CAS number: 69-52-3 )
- Zymolyase®, 20T (Nacalai Tesque, catalog number: 07663-91 )
- FavorPrep Plasmid Extraction Mini Kit (Favorgen, catalog number: FADPE 300 )
- DreamTaq Green PCR Master Mix (2x) (Thermo Fisher Scientific, catalog number: K1082 )
- RedSafe Nucleic Acid Staining Solution (iNtRON Biotechnology, catalog number: 21141 )
- Agarose D1 Low EEO (Condalab, catalog number: 8010.01 )
- LB medium (see Recipes)
- MA2 medium (see Recipes)
- SPA medium (see Recipes)
- STM buffer (see Recipes)
- TBE 20x (see Recipes)
- DNA extraction buffer (see Recipes)
- Transformation buffer (see Recipes)
- Potassium phosphate buffer (see Recipes)
Equipment
- Pipettes P2, P20, P200 and P1000 (e.g., Gilson, PIPETMAN ClassicTM, catalog numbers: F144801 , F144056M , F144058M , F144059M )
- Motorized pipette filler (e.g., Eppendorf, catalog number: 4430000018 )
- Sentino Magnetic Filter Funnels (Pall, catalog number: 4273 )
- Sterile glass handle or glass beads
- Erlenmeyer flasks 500 ml (VWR, catalog number: 10536-926 )
- Filtering flask 250 ml (Simax, catalog number: PJH006 )
- Gene-Pulser (Bio-Rad, model: MicroPulser )
- Heat block (e.g., Eppendorf, catalog number: 5355 000.011 )
- Thermal Cycler (e.g., Bio-Rad, catalog number: 170-6700 )
- Microcentrifuge (e.g., Thermo Scientific, catalog number: 10236249 )
- Centrifuge (e.g., Eppendorf, catalog number: 5703000010 , model: 5702R )
- Power Source (VWR, catalog number: 700-0115 , model: 250 V )
- Gel Doc EZ System (Bio-Rad, catalog number: 1708270 )
- Optical microscope
- Vacuum pump
- Microflow vertical laminar flow workstation
- 28 °C incubator
- Shaker incubator 28 °C
- Autoclave
Software
- Image labTM software 6.0.1. (Bio-Rad, catalog number: 1709690)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Muñoz-Fernández, G., Jiménez, A. and Revuelta, J. L. (2020). Genomic Edition of Ashbya gossypii Using One-vector CRISPR/Cas9. Bio-protocol 10(12): e3660. DOI: 10.21769/BioProtoc.3660.
分类
微生物学 > 微生物遗传学 > CRISPR-Cas9
分子生物学 > DNA > DNA 重组
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link