- EN - English
- CN - 中文
Superresolution Microscopy of Drosophila Indirect Flight Muscle Sarcomeres
果蝇间接飞行肌肌节的超分辨显微镜观察
发布: 2020年06月20日第10卷第12期 DOI: 10.21769/BioProtoc.3654 浏览次数: 5393
评审: Imre GáspárGülçin ÇAKAN AKDOĞANNicanor González-Morales
Abstract
Sarcomeres are extremely highly ordered macromolecular assemblies where proper structural organization is an absolute prerequisite to the functionality of these contractile units. Despite the wealth of information collected, the exact spatial arrangement of many of the H-zone and Z-disk proteins remained unknown. Recently, we developed a powerful nanoscopic approach to localize the sarcomeric protein components with a resolution well below the diffraction limit. The ease of sample preparation and the near crystalline structure of the Drosophila flight muscle sarcomeres make them ideally suitable for single molecule localization microscopy and structure averaging. Our approach allowed us to determine the position of dozens of H-zone and Z-disk proteins with a quasi-molecular, ~5-10 nm localization precision. The protocol described below provides an easy and reproducible method to prepare individual myofibrils for dSTORM imaging. In addition, it includes an in-depth description of a custom made and freely available software toolbox to process and quantitatively analyze the raw localization data.
Keywords: Drosophila flight muscles (果蝇飞行机)Background
Structure of the sarcomeres has been studied in details by X-ray crystallography as well as with various EM methods leading to quasi-atomic models of the thin and thick filaments from numerous species. However, although these examinations resulted in a remarkably good understanding of the actin-myosin overlap region, spatial arrangement of the I-band and H-zone complexes remained largely unknown. Recent advances in fluorescent super-resolution microscopy (also called nanoscopy) provide spatial resolutions that are well below the diffraction limit. Most notably, single-molecule localization microscopy (SMLM) can deliver localization maps of multiprotein complexes with very high precision, virtually attaining a single protein size resolution (Sigal et al. , 2018).
We recently developed a standardized, powerful nanoscopic approach, combined with a structure averaging algorithm, that allowed us to determine the position of 27 muscle proteins in 1 day old adult Drosophila indirect flight muscles (IFMs) with a quasi-molecular, ~5-10 nm localization precision. By using this protein localization atlas, and by means of template based protein structure modeling, we assembled a refined I-band and H-zone model with an unparalleled scope and resolution (Szikora et al. , 2020). For our approach individual myofibrils were isolated from the IFM of Drosophila as described previously (Burkart et al. , 2007; Weitkunat and Schnorrer, 2014), with minor modifications. Individual muscle fibers of the flight muscle are ideal candidates for SMLM because they exhibit an extremely regular structural organization; their intrinsic auto-fluorescence is negligible, and they can be isolated very easily in large numbers. In addition, thickness of the fibers is below ~1.5 µm, therefore the fluorophores can be easily pumped into their dark, non-fluorescent state. As the focal plane is parallel to the myofibrils we can acquire two-dimensional projections of the sarcomeres which reveal the lateral distribution of proteins at the H-zones and I-bands. Since highly specific antibodies are available against different components of the sarcomere, and the organic fluorophores exhibit certain advantageous characteristics, i.e. , high brightness and photostability, we recommend the use of conventional immunofluorescent labeling. Typically, an SMLM measurement generates tens of thousands of microscopic images, to record hundreds of thousands of ‘single molecule events’. To evaluate these large datasets we developed a standardized data-flow protocol. For processing and quantification of the raw localization datasets, we developed a user friendly and freely available software tool (IFM Analyzer) with a detailed user guide including a sample dataset. We trust that with some minor modifications, our approach and the software tools developed can also be used for structural studies of similarly regular or quasi-regular molecular assemblies, such as synaptonemal complexes or chemical synapses.
Materials and Reagents
- Pipette tips
- 6-well plate
- Parafilm
- 1.5 ml microcentrifuge tube
- Precision cover glasses (#1.5, 24 x 24 mm; e.g., Marienfeld-superior; catalog number: 0 117640 )
- Standard fly food media in glass vials (e.g., BDSC Cornmeal Food)
- Cell Culture Multiwell Plate (e.g., Greiner Bio-One, catalog number: 657160 )
- Drosophila (isogenized w1118 strain)
- Glycerol for molecular biology, ≥ 99.0% (Sigma-Aldrich, catalog number: G5516-100ML )
- Adenosine triphosphate (ATP) (Alfa Aesar, catalog number: J10585 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100-500ML )
- Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), molecular biology grade (Merck, catalog number: 324626-25GM )
- Magnesium chloride (MgCl2) (Merck, catalog number: M8266-100G )
- Potassium chloride (KCl) (Merck, catalog number: P9541-500G )
- 1,4-dithiothreitol (DTT) (Merck, catalog number: 10197777001 )
- Phenylmethylsulfonyl fluoride (PMSF) (Merck, catalog number: 10837091001 )
- Paraformaldehyde (Fisher scientific, 16% w/v aq. soln., methanol free, Alfa Aesar, catalog number: 433689M )
- Goat serum (Sigma-Aldrich, catalog number: G9023-10ML )
- Primary antibodies (see Table 1 for examples)
- Secondary antibodies (goat anti-rabbit, anti-mouse or anti-rat IgG highly cross-absorbed antibodies coupled to Alexa647; Thermo Fisher, catalog numbers: A-21245 , A-21235 , A-21247 )
- GFP antibodies (e.g., Thermo Fisher, catalog number: A6455 )
- Nanobodies (e.g., Chromotek, catalog number: gb2AF647 )
- FLAG antibodies (e.g., Sigma-Aldrich, catalog number: F7425 )
- Alexa FluorTM 488 Phalloidin (Thermo Fisher, catalog number: A12379 )
- Glucose (Sigma-Aldrich, catalog number: 49139 )
- Glucose-oxidase (Sigma-Aldrich, catalog number: G2133-50KU )
- Catalase (Sigma-Aldrich, catalog number: C100 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 204439 )
- Tris(hydroxymethyl)aminomethane (Sigma-Aldrich, catalog number: T5941 )
- Tris(2-carboxyethyl)phosphine (TCEP) (Sigma-Aldrich, catalog number: C4706 )
- β-mercaptoethylamine (MEA) (Sigma-Aldrich, catalog number: M6500 )
- Twinsil speed (Picodent, catalog number: 1300 1002 )
- Immersion oil (Olympus Type-F)
- 2x Relaxing solution (see Recipes)
- 1x Relaxing solution (see Recipes)
- Dissection solution (see Recipes)
- Fixative solution (see Recipes)
- Blocking solution (see Recipes)
- dSTORM imaging buffer (see Recipes)
Table 1. Examples of primary antibodies, labeling Drosophila IFM components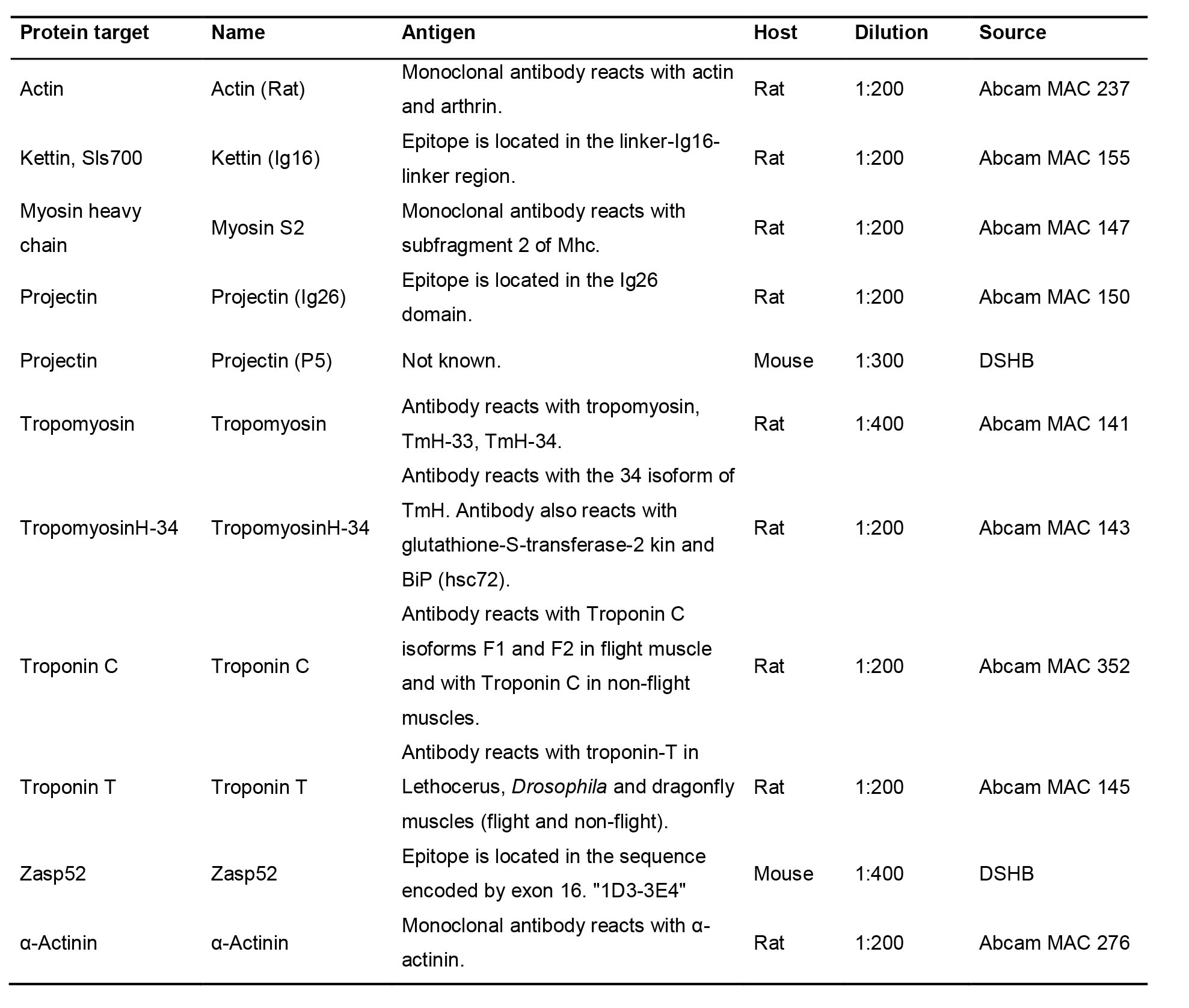
Equipment
- Standard fly husbandry equipment
- CO2 pad (e.g., FlyStuff flypad, Genesee Scientific, catalog number: 59-114 )
- Incubator, set to 25 °C
- Stereo microscope (e.g., Leica microsystems, model: M80 )
- Home-made dissection pad or a commercially available equivalent (see Procedures for details)
- Precision tweezers, straight with fine tips, 0.06 mm (Carl Roth DUMONT, catalog number: K342.1 )
- P2, P20, P100 and P1000 Micro Pipettes (e.g., Gilson, catalog numbers: F144801 , F123600 , F123615 and F123602 )
- Refridgerated centrifuge (e.g., Eppendorf, model: 5424 R , catalog number: 5404000010 )
- Humidity chamber (see Procedures for details)
- 647 nm laser source for fluorescence excitation (647 nm, Pmax = 300 mW; MPB Communications)
- 405 nm laser source for reactivation (405 nm, Pmax = 60 mW; Nichia)
- Nikon LU-N4 laser unit
- Nikon Ti-E inverted microscope frame with oil-immersion objective (CFI Apo 100x, 1.49 NA Nikon)
- Nikon Perfect Focus System
- Quad channel fluorescence filter set (Semrock LF405/488/561/635-A-000, Semrock FF01-446/523/600/677-25)
- Single channel fluorescence filter set (Semrock, catalog numbers: Di03-R635-t1 , BLP01-647R-25 )
- Filter Wheel for emission filters (Thorlabs, catalog number: FW102C )
- Bandpass emission filter 690/70 H (AHF, catalog number: F49-691 )
- 4f optical telescope in detector path, 2* (Thorlabs, catalog number: TTL200 )
- EM-CCD camera (Andor iXon3 897 BV EMCCD)
Software
- NIS Elements AR 5.02 (Nikon)
- Matlab R2018b (Mathworks) or newer with Image Processing Toolbox, Optimization Toolbox, Parallel Computing Toolbox and Curve Fitting Toolbox
- rainSTORM v3.1.7 (available here)
- Minimum System Requirements: 4-core CPU, 32GB RAM
Note: The User Guide is available on the download page. - IFM Analyzer v2.0 (available here)
Note: The User Guide is available on the download page. Example exported section files are available on the download page.
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Szikora, S., Novák, T., Gajdos, T., Erdélyi, M. and Mihály, J. (2020). Superresolution Microscopy of Drosophila Indirect Flight Muscle Sarcomeres. Bio-protocol 10(12): e3654. DOI: 10.21769/BioProtoc.3654.
- Szikora, S., Gajdos, T., Novák, T., Farkas, D., Földi, I., Lenart, P., Erdélyi, M. and Mihály, J. (2020). Nanoscopy reveals the layered organization of the sarcomeric H-zone and I-band complexes. J Cell Biol 219(1). DOI: 10.1083/jcb.201907026.
分类
细胞生物学 > 细胞成像 > 超分辨率成像
分子生物学 > 蛋白质 > 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link
















