- EN - English
- CN - 中文
Safe DNA-extraction Protocol Suitable for Studying Tree-fungus Interactions
适于研究树-真菌互作的安全DNA提取方法
发布: 2020年06月05日第10卷第11期 DOI: 10.21769/BioProtoc.3634 浏览次数: 8026
评审: Amey RedkarDheeraj Singh RathoreAnonymous reviewer(s)
Abstract
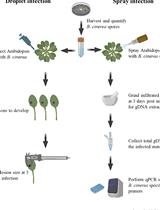
We present a safe and low-cost method suitable for DNA extraction from mycelium and tree tissue samples. After sample preparation, the extraction takes about 60 min. Method performance was tested by extracting DNA from various tree tissue samples and from mycelium grown on solid and liquid media. DNA was extracted from juvenile and mature host material (Picea abies, Populus trichocarpa, Pseudotsuga menziesii) infected with different pathogens (Heterobasidion annosum, Heterobasidion parviporum, Leptographium wagenerii, Sphaerulina musiva). Additionally, DNA was extracted from pure cultures of the pathogens and several endophytic fungi. PCR success rate was 100% for young poplar material and fungal samples, and 48-72% for conifer and mature broadleaved plant samples. We recommend using 10-50 mg of fresh sample for the best results. The method offers a safe and low-cost DNA extraction alternative to study tree-fungus interactions, and is a potential resource for teaching purposes.
Keywords: DNA extraction (DNA 提取)Background
DNA extraction is a central technique in plant-microbe interaction research and for plant disease diagnostic purposes. The abundance of plant-fungal interactions is reflected by the high numbers of cultivable strains isolated from plant samples (Arnold et al., 2001; Higgins et al., 2007; Terhonen et al., 2011). Processing of large numbers of samples for routine PCR reactions with commercial kits can be costly. Additionally, the requirement for hazardous chemicals, such as 2-mercaptoethanol, chloroform, or phenol can restrict the suitability of the protocols for teaching and training purposes.
Due to high secondary metabolite content, many plant samples, and in particular tree tissues, can pose challenges for nucleic acid extraction. Current protocols for DNA extraction from recalcitrant plant tissue utilize organic solvents (chloroform, 2-mercaptoethanol) or surfactants (e.g., cetyl trimethylammonium bromide) (Porebski et al., 1997; Chiong et al., 2017; Yi et al., 2018). Despite their benefits for DNA extraction, these chemicals pose hazards to user health and the environment. Several versions of low-cost, fast, and low health risk protocols for DNA extraction exist for mycelium (Chi et al., 2009), juvenile plant tissue (Edwards et al., 1991; Lu, 2011), grains (Saini et al., 1999), and dried plant tissues (Chabi Sika et al., 2015). However, these methods have not been applied to study tree-fungus interactions, and many times they have been tested only on limited number of sample types. Our goal was to develop and test a safe and low-cost DNA-extraction protocol that is suitable for extracting DNA from various tree tissues and tree-associated fungal samples. After sample preparation, the extraction takes about 60 min. The extracted DNA is suitable for PCR-based downstream applications, such as DNA-based pathogen detection with species-specific primers. Due to its safety and affordability, the method is also a potential resource for teaching and training purposes.
Materials and Reagents
- Standard materials and reagents
- Sterile microcentrifuge tubes, 1.5 or 2.0 ml
- Micropipette tips: 10, 200, 1,000 µl
- Miracloth (Calbiochem, e.g., VWR, catalog number: 475855-1 )
- Plant or fungal samples
- Purified (RO, DI, MilliQ, Nanopure) and sterilized water
- 100 bp DNA ladder (Jena Bioscience, catalog number: M-214S )
- 1 kb DNA ladder (New England Biolabs, catalog number: N0552 )
- NaCl
- KCl
- EDTA
- SDS
- Tris (pH 7.5)
- Polyvinypyrrolidone (PVP, CAS 900-39-8, FW 40,000, e.g., Caisson Labs, catalog number: P071-100GM )
- Isopropanol
- Ethanol (EtOH)
- Extraction buffer (see Recipes)
- Wash buffer (see Recipes)
- Special materials and reagents for different tissue homogenization optionsB.Special materials and reagents for different tissue homogenization options
- Option 1 for soft leaf tissue and mycelium: No special materials or reagents
- Option 2 for various plant tissue and mycelium, larger than 100 mg: Liquid nitrogen
- Option 3 for various plant tissue and mycelium, smaller than 100 mg: Bead beater tubes (e.g., Lysing Matrix I, MP Biomedicals, catalog number: 116918050-CF )
Note: To reduce plastic waste and save on costs, the bead beater tubes can be washed and re-used (see Notes).
Equipment
- Standard equipment
- Scalpels
- Tweezers
- Spatulas
- Scale (e.g., Metler-Toledo, model: ML54T )
- Micropipettes: 1, 10, 100, 1,000 µl
- Heat block (e.g., VWR, catalog number: 12621-096 )
- Vortex (e.g., VWR, catalog number: 10153-838 )
- Microcentrifuge (e.g., Eppendorf, model: 5424 )
- One of the following to heat up water: Microwave, waterbath (e.g., VWR, model: WB05 ), or hot plate (e.g., VWR, catalog number: NO97042-642 )
- Freezer, -20 °C or -80 °C
- Special equipment for different tissue homogenization options
- Option 1 for soft leaf tissue and mycelium: No special equipment
- Option 2 for various plant tissue and mycelium, more than 100 mg:
Dewar for liquid nitrogen
Ceramic mortars and pestles - Option 3 for various plant tissue and mycelium, less than 100 mg:
Bead beater (e.g., Biospec, model: Mini-Beadbeater 16 , catalog number: 607/607EUR ) - For sampling xylem tissue from mature trees: chisel (e.g., Grainger, catalog number: 2AJA6 ) and mallet (e.g., Grainger, catalog number: 4YR61 ), cutting board
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Keriö, S., Terhonen, E. and LeBoldus, J. M. (2020). Safe DNA-extraction Protocol Suitable for Studying Tree-fungus Interactions. Bio-protocol 10(11): e3634. DOI: 10.21769/BioProtoc.3634.
分类
植物科学 > 植物分子生物学 > DNA > DNA 提取
微生物学 > 病原体检测 > PCR
分子生物学 > DNA > DNA 提取
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link