- EN - English
- CN - 中文
Evaluation of the Efficiency of Genome Editing Tools by a Frameshift Fluorescence Protein Reporter
通过移码荧光蛋白报告基因评估基因组编辑工具的效率
(*contributed equally to this work) 发布: 2020年05月20日第10卷第10期 DOI: 10.21769/BioProtoc.3622 浏览次数: 5091
评审: Alba BlesaShyam SolankiChangyi Zhang
Abstract
In the last decade, genome editing has been the center of attention as a novel tool for mechanistic investigations and for potential clinical applications. Various genome editing tools like meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector-based nucleases (TALEN), and the clustered regularly interspaced short palindromic repeats (CRISPR)-associated genes (Cas), have been developed in recent years. For the optimal use as well as continued developments of these genome editing tools, the evaluation of their efficiencies and accuracies is vital. Here, we present a protocol for a reporter based on frameshift fluorescence protein which we recently developed to evaluate the efficiency and accuracy of genome editing tools. In this method, a ~20 bp target sequence containing frame-shifting is inserted after the start codon of a cerulean fluorescence protein (CFP) to inactivate its fluorescence, and only a new insertion/deletion event in the target sequence will reactivate the CFP fluorescence. To increase the traceability, an internal ribosome entry site and a red fluorescence protein, mCherryFP, are placed downstream of the reporter. The percentage of CFP-positive cells resulted from in/del mediated fluorescence restoration can be quantified by fluorescence measuring devices as the readout for genome editing frequency. As a demonstration, we present the usage for CRISPR-Cas9 technique here with flow cytometer as the readout for fluorescence changes.
Keywords: Insertion-deletion (碱基插入缺失)Background
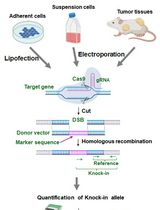
Genome editing tools are very important for the investigations of biological mechanisms and prevention and/or treatment of genetic diseases (Maeder and Gersbach, 2016). In the last couple of decades, several genome editing tools have been introduced, which include the meganucleases (Epinat et al., 2003), zinc finger nucleases (ZFNs) (Kim et al., 1996), transcription activator-like effector-based nucleases (TALEN) (Christian et al., 2010), and the clustered regularly interspaced short palindromic repeats (CRISPR)-associated genes (Cas) (Jinek et al., 2012; Cong et al., 2013; Sander and Joung, 2014). In general, these tools create DNA double stranded breaks (DSB) to trigger genome editing in vivo (Maeder and Gersbach, 2016). The evaluation of the efficiencies and specificities of genome editing tool is essential for their applications and further developments. In our recent published study, we described a reporter that can generate quantitative readout for genome editing efficiency (Kumar et al., 2019). In this system, a ~20 bp target sequence is placed in a multiple cloning site (MCS) which is right after the start codon of Cerulean fluorescence protein (CFP) to generate a frame-shift of the open reading frame (ORF). This frameshifted-CFP (FsCFP) can be used as a reporter of genome editing because only when there is a successful DNA-double strand break (DSB) event on the target sequence followed with a non-homologous end joining (NHEJ) to generate an in/del event to shift the reading frame to a correct order (by a chance of up to 1/3), the CFP fluorescence will be reactivated as a positive readout. To facilitate the quantification, an internal ribosome entry site (IRES) and a red fluorescence protein, mCherryFP, is placed after the reporter. In principle, this reporter can be applied to any genome editing system as long as a DSB and NHEJ are expected from the editing. This approach can effectively detect low-efficiency editing in a population of cells with very low false negative or false positive. Furthermore, in this method, the positive cells can be conveniently identified and enriched for the examination or validation of the in/del event in the genome. Also, this method can be easily adapted for screening to optimize the genome-editing enzyme or the other components (such as guide-RNA) in the positive cells. Here, we used the CRISPR-Cas9 technique as a demonstration and the flow cytometry as the readout of the fluorescence events.
In this protocol, the target sequence is inserted between restriction sites of NotI and XhoI before CFP reporter together with sequence for the optimal recognition of Cas9 and a premature STOP codon to create a frame shift. The reporter region is then integrated in the nuclear genome of the target cell by the assistance of lentivirus. The target cells expressing the red fluorescence protein are then isolated by fluorescence-activated cell sorting (FACS), before vectors containing the Cas9 and gRNA are introduced into these cells. After incubation, the ratio of the CFP over mCherryFP was measured in flow cytometry to provide quantitative measurement for the efficiency of the genome editing.
Materials and Reagents
Materials
- Cell Culture dish 150 x 25 mm (Asi, catalog number: TD0150 )
- Cell Culture dish 90 x 20 mm (Asi, catalog number: TD0100 )
- 5 ml serological pipet (Asi, catalog number: SP205 )
- 10 ml serological pipet (Asi, catalog number: SP210 )
- Cell Culture Flask 75 cm2, filter cap (Asi, catalog number: TV0075 )
- Cell Culture Flask 25 cm2, filter cap (Asi, catalog number: TV0025 )
- Syringe filter, PES 25 mm, 0.45 μm (Asi, catalog number: TE45-5 )
- 10 ml syringes (BD, catalog number: 309604 )
- 15 ml and 50 ml conical tubes (Denville, catalog number: C1062-P )
- 18 G x 1 ½ needles (BD, catalog number: 305196 )
- Bottle top filtration–2 μm PES (VWR, catalog number: 97066-202 )
- 15 ml centrifuge tubes (CellPro, catalog number: CN5600 )
- 50 ml centrifuge tubes (CellPro, catalog number: CN5603 )
- Filter Pipet Tips 1,250 μl (TruPoint, catalog number: FT1250 )
- Filter Pipet Tips 200 μl (TruPoint, catalog number: FT1200 )
- Filter Pipet Tips 20 μl (TruPoint, catalog number: FT1020 )
- Filter Pipet Tips 10 μl (TruPoint, catalog number: FT1010 )
- Falcon FACS tubes with 35 μMcell strainer (BD, catalog number: 352235 )
- Falcon FACS collection tubes (BD, catalog number: 352063 )
- Ice buckets
- T4 DNA ligase (New England Biolabs, catalog number: M0202S )
- Gel extraction kit (EZ, catalog number: M1002-50 )
- One ShotTM TOP10 Chemically Competent E. coli (Invitrogen, catalog number: C404003 )
- ZymoPURETM II Plasmid Midiprep Kit (Genesee Scientific, catalog number: 11-550B )
- QIAprep Spin Miniprep Kit (50) (QIAGEN, catalog number: 27104 )
Cells and Plasmids
- Human embryonic kidney cells (HEK 293T, clone 17) (ATCC, catalog number: CRL-11268 )
- Plasmid pQC-XIG (Addgene, catalog number: w497-1 ), deposited by Dr. Eric Campeau, who is currently at Zenith Epigenetics Ltd, Calgary, Canada
- Plasmid pCMV-Delta R8.2 (Addgene, catalog number: 12263 ), deposited by Dr. Didier Trono at EPFL
- Plasmid pCMV-VSV-G (Addgene, catalog number: 8454 ), deposited by Dr. Robert Weinberg at MIT
- gRNAs custom ordered from Vector builder (https://en.vectorbuilder.com/)
Reagents
- High Glucose DMEM (1x) (Life Technologies, catalog number: 11995-065 )
- F-10 media (1x) (Life Technologies, catalog number: 11550-043 )
- 0.25% Trypsin-EDTA (1x) (Life technologies, catalog number: 25200-072 )
- 10% FBS (Hyclone, catalog number: SH30910.03 )
- Polybrene Transfection Reagent (Millipore, catalog number: TR-1003-G )
- Antibiotic-Antimycotic (100x) (Life Technologies, catalog number: 15240062 )
- Polyethylenimine (PEI) (Sigma-Aldrich, catalog number: 408727 )
- T4-ligase Buffer (New England Biolabs, containing 50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 10 mM DTT, pH 7.5)
- DMEM+F10 culture media (45% DMEM + 45% F-10 + 10% FBS, with 1x Antibiotic-Antimycotic)
- Nucleotide oligos/primers:
Table 1. List of primers used in this protocol
Equipment
- Centrifuge 5424 R (Eppendorf, catalog number: 540400138 )
- Labnet Accublock Digital Dry Bath (Labnet, catalog number: 19-41620 )
- Gene Mote Vortex Mixer (Bioexpress, catalog number: S-3200-1 )
- CO2 incubator MCO-19AIC (UV) (Panasonic, catalog number: 13010002 )
- Centrifuge 5702 (Eppendorf, catalog number: 0 22626205 )
- Fluorescent microscope
- Aria-IIU flow cytometer (BD)
Software
- FCS Express 6 (Denovo software–https://denovosoftware.com/)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Moorthy, B. T., Kumar, A., Lotenfoe, L. X. and Zhang, F. (2020). Evaluation of the Efficiency of Genome Editing Tools by a Frameshift Fluorescence Protein Reporter. Bio-protocol 10(10): e3622. DOI: 10.21769/BioProtoc.3622.
分类
生物化学 > 蛋白质 > 荧光
癌症生物学 > 基因组不稳定性及突变 > 细胞生物学试验 > DNA结构和改变
分子生物学 > DNA > DNA 损伤和修复
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link