- EN - English
- CN - 中文
Negative Ion Mode nanoLC-ESI-MS/MS Analyses of Permethylated Sulfated Glycans
甲基化硫酸多糖的负离子模式nanoLC-ESI-MS / MS分析
发布: 2020年05月20日第10卷第10期 DOI: 10.21769/BioProtoc.3618 浏览次数: 4359
评审: Manjula MummadisettiAnonymous reviewer(s)
Abstract
We have developed enabling techniques for sulfoglycomics based on MALDI-MS mapping and MS/MS sequencing of permethylated sulfated glycans. We then extended further the analytical workflow to C18 reverse phase (RP)-nanoLC-nanoESI-MS/MS analyses of permethylated sulfated glycans in the negative ion mode. The advantages are that extra sulfates on permethylated di- and multiply sulfated glycans will survive in nanoESI conditions to allow detection of multiply charged intact molecular ions, and more comprehensive MS/MS can be performed in an automated fashion at higher sensitivity, compared with MALDI-MS/MS. Parallel higher energy collision dissociation (HCD) and ion trap collision induced dissociation (CID)-based MS2, coupled with product-dependent MS3 in data dependent acquisition mode proved to be highly productive when applied to resolve and identify the isomeric sulfated glycan structures. In-house glycomic data mining software, GlyPick, was developed and used to automate the downstream process of identification and relative quantification of target sulfated glycotopes based on summed intensity of their diagnostic MS2 ions extracted from thousands of HCD-MS2 and/or CID-MS2 data.
Keywords: Permethylated sulfated glycans (全甲基化硫酸多糖)Background
Current mass spectrometry (MS)-based glycomic mapping remains insufficient to delineate the full complexity of the glycome. Although MALDI-MS mapping and MS/MS sequencing would efficiently afford a very useful first impression glycomic profile, it is nearly impossible to acquire MALDI-MS/MS on every putative glycan signal detected, especially those of low intensities and/or occurring at higher masses. In that respect, LC-ESI-MS/MS analysis in an automated and data-dependent acquisition (DDA) mode provides a far more comprehensive MS/MS data coverage (Cheng et al., 2013; Cheng et al., 2015; Hsiao et al., 2017; Yu et al., 2018). We have demonstrated that permethylation in conjunction with 2 steps C18/amine SPE or a single step mixed mode MAX SPE fractionation can yield fully methylated non-sulfated, mono-sulfated, and di-sulfated glycans in separate pools (Yu et al., 2020) for MALDI-MS screening. We found that loss of sodium sulfite from permethylated di-sulfated glycans occurred readily during MALDI-ionization (Figure 1A), while the two sulfates on permethylated glycans were fully retained when analyzed by nanoESI-MS in negative ion mode, allowing them to be detected as [M-2H]2- (Figure 1B), and further selected for MS/MS analyses at high sensitivity. Moreover, we showed that the negative mode nanoLC-MS/MS analysis of permethylated sulfated glycans on a C18 reverse phase capillary column could be efficiently carried out using the common acetonitrile/formic acid/water solvent system, and hence be fully compatible with the normal set-up of an analytical laboratory devoted to proteomics.
By virtue of a panel of synthetic sulfated glycans, we have shown that the beam-type higher energy collision dissociation (HCD) MS/MS as implemented on the hybrid Orbitrap series would afford a range of low mass fragment ions. These diagnostic ions would define the location of sulfate on which glycosyl residue at which position (Cheng et al., 2015). Ion trap-based CID MS2, on the other hand, would not retain these very useful ions due to its one third low mass cut-off (Patnode et al., 2013). It does, however, offer a higher sensitivity and acquisition rate, allowing definitive assignment of target sulfated glycotopes via MS2 product ion-dependent MS3. Occasionally, when a di-sulfated O-glycan may carry a different permutation of sulfated glycotopes on either the 6-arm or the extended 3-arm, additional targeted MSn analyses can help identify the existence of isomers (Yu et al., 2018). Moving from the original linear ion trap-Orbitrap hybrid systems to more recent tribrid Orbitrap Fusion systems, the current MS systems afford a higher degree of flexibility for different combination of single to multiple stages of HCD versus ion trap-based CID MS/MS to be acquired either in Orbitrap for greater resolution and mass accuracy, or the ion trap for better sensitivity and speed. These aspects will not be further dealt with here. Suffice to point out that the basic principle remains the same, namely to acquire as many HCD/CID MS2 within an elution time window-compatible DDA duty cycle and to couple each, if possible and desirable, to a pre-determined list of product-dependent MS3 for greater depth of structural details. With or without MS3, it is important that the low mass ions produced in negative mode MS2 be retained and detected at reasonably high resolution and mass accuracy (< 5 ppm, if possible).
Online LC and data-dependent MS/MS acquisition will produce a huge dataset which is near impossible to manually analyze systematically. Unlike proteomics, neither the glycan database nor the MS/MS sequencing algorithm is well developed to allow direct MS2 ions or spectral matching search for unambiguous identification. An in-house computational tool, GlyPick, was developed to filter out bona fide glycan MS2 spectra by user-defined criteria, usually by presence of at least 2 to 3 diagnostic glycan fragment ions. It can also extract out and compute the occurrence and summed intensity of a list of user-input diagnostic MS2 ions that will define the presence and relative abundance of those important glycotopes such as 6-sulfated GlcNAc, 3’-sulfated Gal, 6’-sulfated Gal, sulfated LacNAc, sulfated Le, etc, from these MS2 spectra (Hsiao et al., 2017; Yu et al., 2018). Results are output in CSV format, which can be conveniently interrogated using Excel for further data mining and graph plotting.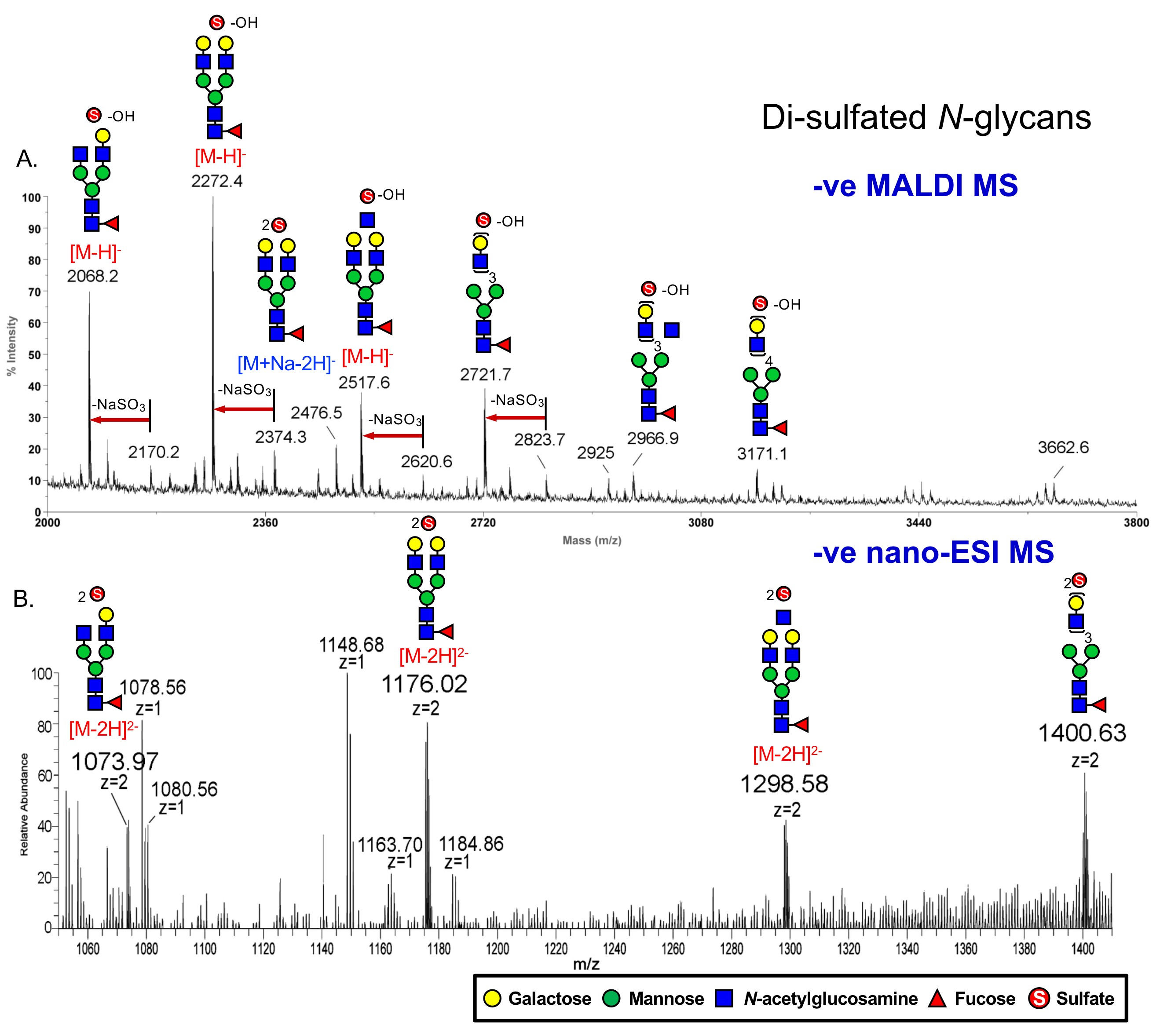
Figure 1. MS spectra of di-sulfated permethylated N-glycans acquired by different ionizations, which are (A) MALDI-MS and (B) nanospray ESI-MS, both in the negative ion mode.
Materials and Reagents
- Plastic pipette tips
- Microcentrifuge tube
- ZipTipC18 (Merck Millipore, catalog number: ZTC18S096 )
- Acetonitrile with 0.1% (v/v) Formic Acid for LC-MS, JT Baker® (VWR, catalog number: JT9832-2 )
- Water with 0.1% (v/v) Formic acid, BAKER ANALYZEDTM, JT Baker® (VWR, catalog number: JT9826-3 )
Equipment
- SpeedVac
- nanoACQUITY UPLC system (Waters Corporation)
- nanoACQUITY M-Class BEH130 C18 column, 1.7 μm, 75 μm x 250 mm (Waters Corporation, catalog number: 186003545 )
- Picoview nanospray source 550 (New Objective, catalog number: PV-550 )
- A hybrid LTQ-Orbitrap EliteTM Mass Spectrometer (Thermo Scientific), or any equivalent high resolution/mass accuracy MS system including the tribrid Orbitrap Fusion (Thermo Fisher Scientific)
Software
- MasslynxTM software v. 4.1 (Waters)
- XcaliburTM software v2.2 (Thermo Fisher Scientific)
- GlyPick (in-house, available upon request)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yu, S., Cheng, C. and Khoo, K. (2020). Negative Ion Mode nanoLC-ESI-MS/MS Analyses of Permethylated Sulfated Glycans. Bio-protocol 10(10): e3618. DOI: 10.21769/BioProtoc.3618.
- Yu, S. Y., Hsiao, C. T., Izawa, M., Yusa, A., Ishida, H., Nakamura, S., Yagi, H., Kannagi, R. and Khoo, K. H. (2018). Distinct substrate specificities of human GlcNAc-6-sulfotransferases revealed by mass spectrometry-based sulfoglycomic analysis. J Biol Chem 293(39): 15163-15177.
分类
生物化学 > 糖类 > 多糖
生物化学 > 糖类 > 糖蛋白
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link