- EN - English
- CN - 中文
Rapid Generation of Human Neuronal Cell Models Enabling Inducible Expression of Proteins-of-interest for Functional Studies
快速建立能诱导表达功能研究所需蛋白的人神经细胞模型
发布: 2020年05月05日第10卷第9期 DOI: 10.21769/BioProtoc.3615 浏览次数: 5301
评审: Ehsan KheradpezhouhNarayan SubramanianAnonymous reviewer(s)

相关实验方案
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1216 阅读
Abstract
CRISPR-Cas9 technology has transformed the ability to edit genomic sequences and control gene expression with unprecedented ease and scale. However, precise genomic insertions of coding sequences using this technology remain time-consuming and inefficient because they require introducing adjacent single-strand cuts through Cas9 nickase action and invoking the host-encoded homology-directed repair program through the concomitant introduction of large repair templates. Here, we present a system for the rapid study of any protein-of-interest in two neuronal cell models following its inducible expression from the human AAVS1 safe harbor locus. With lox-flanked foundation cassettes in the AAVS1 site and a tailor-made plasmid for accepting coding sequences-of-interest in place, the system allows investigators to produce their own neuronal cell models for the inducible expression of any coding sequence in less than a month. Due to the availability of preinserted enhanced green fluorescent protein (EGFP) coding sequences that can be fused to the protein-of-interest, the system facilitates functional investigations that track a protein-of-interest by live-cell microscopy as well as interactome analyses that capitalize on the availability of exquisitely efficient EGFP capture matrices.
Keywords: Human neuronal cells (人神经元细胞)Background
The ability to engineer the genomes of cell models and organisms holds tremendous potential for research and targeted therapy. With the advent of CRISPR-Cas9 technology, the efficiency of precision genome engineering has greatly improved, and useful reagents are coming online at a rapid pace. Despite these advances, there still is a scarcity of cell-based in vitro paradigms for studying neurological diseases that is holding back the pace of progress in this area. Whereas CRISPR-Cas9 mediated homology-directed repair (HDR) can now easily be applied to generate knockout models or to knock in small transgenic segments, the goal to integrate larger segments continues to present a considerable challenge (Devkota, 2018).
In our own work, we often would like to learn more about the function of a protein by studying its subcellular localization and protein-protein interactions. To this end, it would help if neuronal cell models could be equipped with inducible expression systems that provide temporal control of the expression of a protein-of-interest and facilitate its visualization and capture. Not long ago, this need would call for the stable transfection or lentiviral transduction of an expression cassette, an approach that can result in unpredictable confounding effects due to the random integration of transgenes. CRISPR-Cas9 provides alternative strategies to accomplish this task in a precise way but these approaches remain time-consuming and require considerable investments in resources and expertise.
To address this unmet need, we sought to develop resources that enable the rapid and flexible integration of large inducible expression cassettes into the AAVS1 human safe harbor locus (DeKelver et al., 2010) of several neuronal cell models. Stable insertion into the AAVS1 locus, as opposed to viral integration, was chosen to preclude transgene silencing and insertional mutagenesis, notorious confounders associated with non-directed transgene insertions. To maximize flexibility and speed, the system was built around two genome-editing steps. The first step, which makes use of CRISPR-Cas9-mediated HDR followed by selection of positive clones, is time-consuming and uncertain to succeed but we have already accomplished this in 3 cell lines that are available to be shared. The second step, which relies on a Cre recombinase-mediated exchange of an expression cassette, is relatively easy to accomplish and fast because it can be followed by puromycin-based selection of positive clones.
More specifically, a paired Cas9 nickase CRISPR strategy, used to improve the specificity of targeting and to minimize off-target effects (Ran et al., 2013), was employed to insert a foundation cassette (FC), comprising a pair of lox sites flanking a G418 resistance marker into Intron 1 of the AAVS1 locus, a human genetic safe harbour. In parallel, a ~7 kb inducible expression plasmid (IEX) was built. The expression cassette within the IEX is flanked by compatible lox sites and comprises elements designed to accept the insertion and control the expression of any coding sequence-of-interest (Figure 1). More specifically, the IEX comprises a promoter-less puromycin resistance marker, the reverse transactivator (rtTA3), and an EGFP tag fused to a protein-of-interest insertion site. With this arrangement, the rtTA3 is designed to drive the inducible expression of the EGFP-fused protein-of-interest that is under the control of the TREtight inducible promoter.
With these tools in hand, novel cell models for the inducible expression of a protein-of-interest fused to EGFP can be generated in three straightforward steps:
1.Insert coding sequence for protein-of-interest into the IEX plasmid.
2.Transiently co-transfect the IEX plasmid and a plasmid coding for Cre recombinase into the parental cell equipped with one or two FCs.
3.Select stable integrant clones by puromycin selection or fluorescence activated cell sorting (FACS).
In addition to its ease of handling and speed, the system restricts the copy number of insertions to two, corresponding to the two AAVS1 alleles present in the human genome. The system also lends itself to comparative analyses of several proteins, because its design ensures that cells coding for different proteins-of-interest can be generated side-by-side, requiring no specific resources, except for a human cDNA library and the corresponding primers to amplify and insert a given coding sequence into the IEX cassette. Note that fusing the coding sequence for a protein-of-interest to EGFP does not only provide the ability to undertake live cell imaging but the EGFP tag also allows the high affinity purification of fusion proteins by GFP nanotrap technology for the study of protein-protein interactions (Rothbauer et al., 2008).
We recently applied the system to study the TAU protein and BAG5 in several human neuronal cell models, including ReN VM human neural progenitor cells that can be differentiated into a co-culture of neural and glial cells (Donato et al., 2007). These studies have led us to detect subtle differences in protein-protein interactions of wild-type versus mutant forms of TAU and BAG5 (Figure 1). This article was written with the intent to share these resources and facilitate their adaptation to the study of other proteins-of-interest.
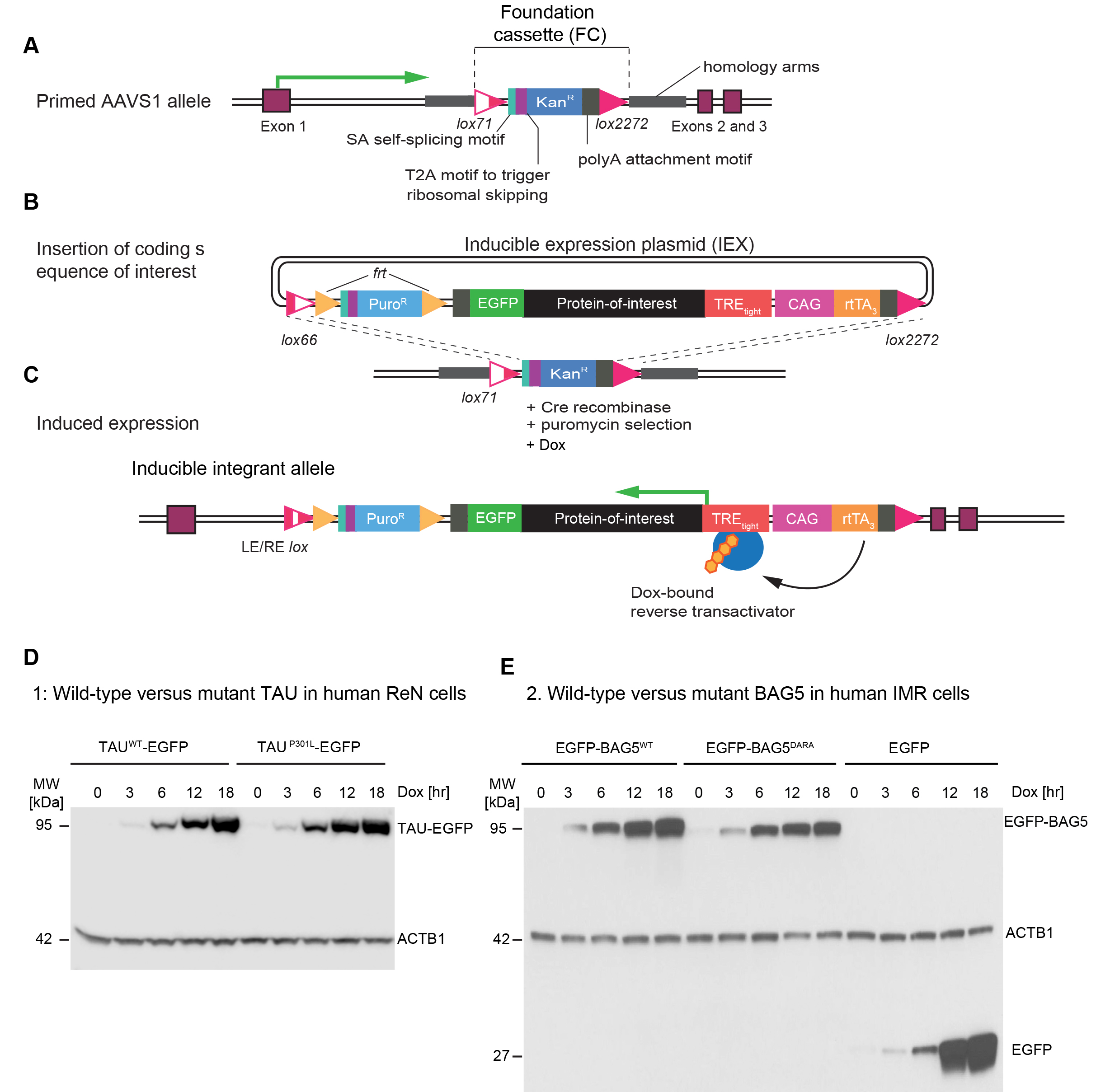
Figure 1. Design of AAVS1 embedded system for inducible expression of proteins-of-interest. A. CRISPR-Cas9 nickase-mediated insertion of foundation cassette (FC) into AAVS1 safe harbour. B. Insertion of coding sequence-of-interest into inducible expression plasmid (IEX). C. Reverse-tetracycline-controlled transactivator (rtTA3)-mediated expression of protein-of-interest following addition of doxycycline to cell culture medium. D. Example 1: Parallel induction of wild-type and P301L mutant TAU fused to C-terminal EGFP in ReN cells. Antibodies directed against beta-actin served as the loading control. E. Example 2: Side-by-side induction of N-terminally EGFP-tagged wild-type or mutant (DARA) BAG5. Cells expressing EGFP only served as additional controls in this analysis.
Materials and Reagents
- Biological materials
- Plasmids (Table 1)
- Cell lines (Table 2)
Table 1. List of available plasmids for generating inducible cells and their Addgene ID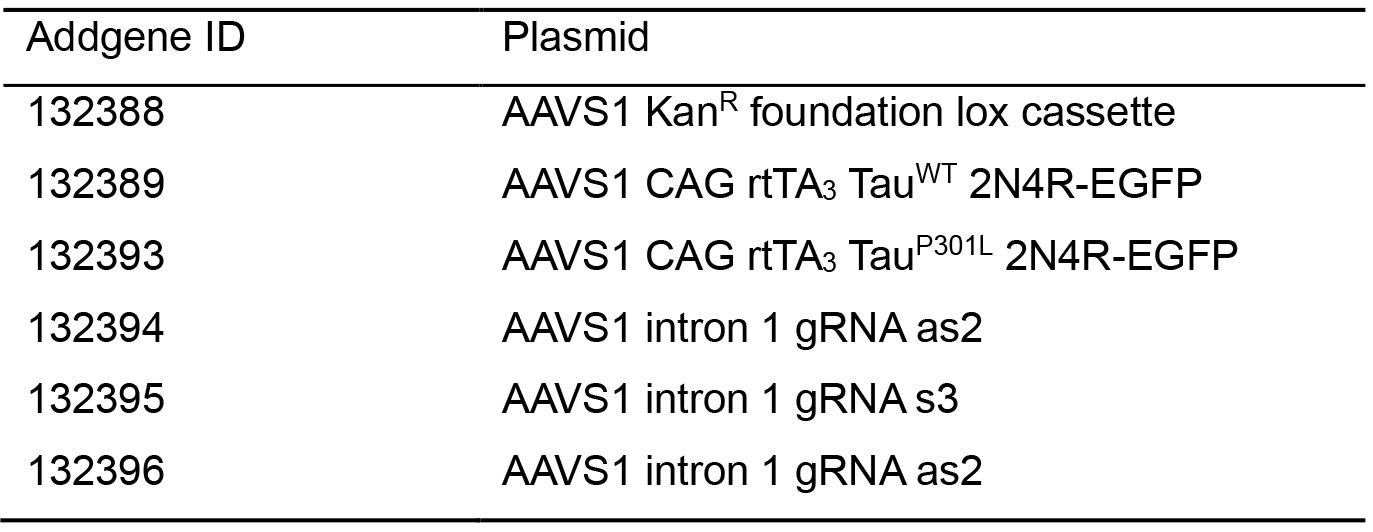
Table 2. List of available cell lines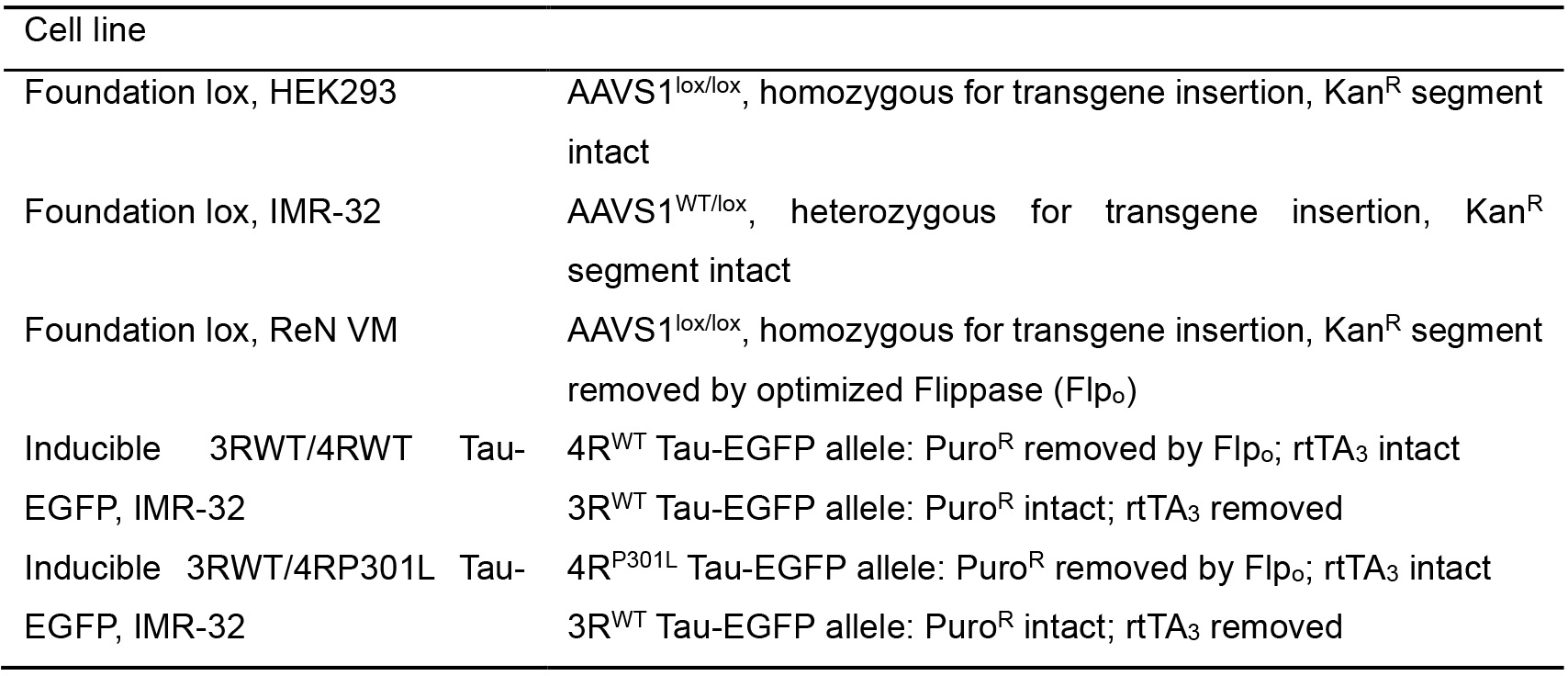
- Cloning
Note: All cloning enzymes and PCR reagents are stored at -20 °C, stable for 1 year.- Q5® Hot Start high-fidelity 2x master mix (New England Biolabs, catalog number: M0494S )
- Primers: ordered as Value Oligos or Custom Oligos, 25 nmole, desalted (Invitrogen). Reconstitute to 100 μM stocks in water, store at -20 °C
- Q5® Site-directed mutagenesis kit (New England Biolabs, catalog number: E0554S )
- EZ-10 spin column PCR products purification kit (BioBasic, catalog number: BS664-250preps )
- Restriction enzyme BbvCI with CutSmart buffer (New England Biolabs, catalog number: R0601S )
- Restriction enzyme NheI-HF with CutSmart buffer (New England Biolabs, CutSmart, catalog number: R3131S )
- Fast AP thermosensitive alkaline phosphatase (Thermo Fisher Scientific, catalog number: EF0651 )
- PureLink Quick Gel Extraction Kit (Invitrogen, catalog number: K210012 )
- T4 DNA ligase (New England Biolabs, catalog number: M0202S )
- EnduraTM Chemically Competent E. coli Cells (Lucigen, catalog number: 60241-2 ). Store at
-80 °C - EZ-10 spin column plasmid DNA minipreps kit (BioBasic, catalog number: BS614-250preps )
- PureLink® HiPure plasmid filter maxi kit (Invitrogen, catalog number: K210026 )
- Ampicillin sodium salt powder, 91.0-100.5%, cell culture tested (Sigma-Aldrich, catalog number: A1066-5G ). Store at 4 °C, stable for 1 year
- Inducible cell generation
- paavCAG-iCre plasmid (Addgene, catalog number: 51904 , a gift from Jinhyn Kim)
- jetPRIME (PolyPlus, catalog number: 114-07 ). Store at 4 °C, stable for 1 year
- TransfeX transfection reagent (ATCC, catalog number: ACS-4005 ). Store at 4 °C, stable for 1 year
- Millex-HA syringe filters, 0.45 μm pore size (Millipore, catalog number: 5010-SLHA033SS )
- Millex-GP syringe filters, 0.22 μm pore size (Millipore, catalog number: SLGP033RS )
- 6-well tissue-culture plates (Falcon, catalog number: 353046 )
- 24-well tissue-culture plates (Falcon, catalog number: 353047 )
- Puromycin (Sigma-Aldrich, catalog number: P7255-25MG ). Reconstitute to 10 mg/ml stock, filter sterilize, aliquot, store at -20 °C
- Doxycycline (BioBasic, catalog number: DB0889-25G ). Reconstitute to 10 mg/ml stock, aliquot, store at -20 °C
- Affinity capture with GFP nanotrap
- Cell culture
- 6-cm tissue-culture plates (Sarstedt, catalog number: 83.3901 )
- 1.5 ml centrifuge tubes (Sarstedt, catalog number: 72.690.300 )
- 12-well tissue-culture plates (Sarstedt, catalog number: 83.3921 )
- Dulbecco's Phosphate-Buffered Saline (D-PBS), without calcium chloride and magnesium chloride, liquidPhosphate buffered saline (Sigma-Aldrich, catalog number: D8537-500ML ). Store at 4 °C, stable for 1 year5.
- 0.25% Trypsin with EDTA 4Na (Gibco, catalog number: 25200072 ). Aliquot, store at -20 °C for long-term storage. Store at 4 °C for short-term use, stable for 6 months
- StemPro® Accutase® Cell Dissociation Reagent (Gibco, catalog number: A1110501 ). Aliquot, store at -20 °C for long-term storage. Store at 4 °C for short-term use, stable for 6 months
- Matrigel, growth-factor reduced (Corning, catalog number: 354230 ). Aliquot, store at -20 °C, stable for 1 year
- RecoveryTM Cell Culture Freezing Medium (Gibco, catalog number: 12648010 ). Aliquot, store at -20 °C, stable for 1 year
- DMEM with high glucose, L-glutamine, and sodium pyruvate (Gibco, catalog number: 11995073 ). Store at 4 °C, stable for 1 year
- 10% Fetal Bovine Serum, qualified (Gibco, catalog number: 12483020 ). Aliquot, store at -20 °C for long-term storage
- 1% GlutaMAXTM-I supplement (Gibco, catalog number: 35050061 ). Store at 4 °C, for stable for 1 year
- DMEM/F12, 1:1, contains L-glutamine, but no HEPES (Gibco, catalog number: 21041025 ). Store at 4 °C, stable for 1 year
- 2% N21-MAX supplement (R&D Systems, catalog number AR008 ) or 2% B-27 serum-free supplement (Gibco, catalog number: 17504044 ). Store at -20 °C, stable for 1 year
- Basic fibroblast growth factor (Gibco, catalog number: PHG0261 ). Store at -80 °C, stable for 1 year
- Epidermal growth factor (Reprokine, catalog number: RKP01133 ). Store at -80 °C, stable for 1 year
- Heparin (Sigma-Aldrich, catalog number: H3149-10KU ). Store at -80 °C, stable for 1 year
- 2 ng/ml glial-derived neurotrophic factor (Gibco, catalog number: PHC7045 )
- 500 μM dibutyryl-cyclic-adenosine monophosphate (Sigma-Aldrich, catalog number: D0627-250MG )
- Proliferation media for IMR-32 and HEK-293 cells (see Recipes)
- Proliferation media for ReN VM cells (see Recipes)
- Neuronal differentiation media for ReN VM cells (see Recipes)
- Cell lysis
Note: Original reagents are stored at room temperature. Reconstituted stocks are stored at 4 °C for 3 months. All buffers (see Recipes) are prepared fresh from reagent stocks immediately before use.- Tris-HCl, > 99% (BioShop, catalog number: TRS002.5 ). Reconstitute to 1 M stock in water
- NP-40 (BioShop, catalog number: NON505.500 ). Reconstitute to 10% stock in water
- Sodium deoxycholate (DOC), > 99% (BioShop, catalog number: DCA333.50 ). Reconstitute to 10% w/v stock in water, pH 8.0
- Sodium chloride (NaCl), > 99% (BioShop, catalog number: SOD002.205 ). Reconstitute to 5 M stock in water
- Ethylenediaminetetraacetate (EDTA), >99% (BioShop, catalog number: EDT001.500 ). Reconstitute to 0.5 M stock in water
- Sodium orthovanadate (Na3VO4) (BioShop, catalog number: SOV664.25 ). Reconstitute to 0.1 M stock in water
- Sodium fluoride (NaF) (BioShop, catalog number: SFL001.100 ). Reconstitute to 1 M stock in water
- Phenylmethylsulfonyl fluoride (PMSF) (BioShop, catalog number: PMS123.5 ). Reconstitute to 0.5 M stock in ethanol
- cOmplete protease inhibitor (Roche, catalog number: 11836170001 )
- PhosStop phosphatase inhibitor (Roche, catalog number: 4906837001 )
- HEPES (Bioshop, catalog number: HEP001.500 ). Reconstitute to 0.1 M stock in water
- Trifluoroacetic acid (TFA), > 99%, HPLC grade (Sigma-Aldrich, catalog number: 302031-100ML )
- Acetonitrile (ACN), HPLC grade (Caledon, catalog number: 1401-7-40 )
- Lysis buffer (see Recipes)
- Wash buffer (see Recipes)
- Elution buffer (see Recipes)
Equipment
- -80 °C freezer (Thermo Fisher, model: Standard Performance Ultra-Low Freezers )
- Refrigerated micro centrifuge (Eppendorf, model: 5424R , catalog number: 5404000138 )
- Avanti J-26S series high-speed centrifuge (Beckman-Coulter, catalog number: B22984 )
- Orbital shaker (Jeio Tech, model: ISF-7000 series )
- Fluorescence microscope (Leica Microsystems, model: Leica DMI 6000 B )
- NanoDrop spectrophotometer (Thermo Fischer Scientific, catalog number: ND-1000 )
- ESBE Scientific Save cell UV cell culture incubator (Panasonic, catalog number: MCO-19AIC )
- SterilGARD biosafety cabinet (The Baker Company, catalog number: SG603A-HE )
Software
- Snapgene (GSL Biotech LLC, https://www.snapgene.com/)
- Snapgene Viewer (GSL Biotech LLC, https://www.snapgene.com/snapgene-viewer/)
- Tm Calculator (New England Biolabs, https://tmcalculator.neb.com/#!/main)
- NEBioCalculator (New England Biolabs, https://nebiocalculator.neb.com/#!/ligation)
- NanoDrop 1000 (Thermo Fischer Scientific, https://www.thermofisher.com/ca/en/home/industrial/spectroscopy-elemental-isotope-analysis/molecular-spectroscopy/ultraviolet-visible-visible-spectrophotometry-uv-vis-vis/uv-vis-vis-instruments/nanodrop-microvolume-spectrophotometers/nanodrop-software-download.html)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Wang, X., Friesen, E., Müller, I., Lemieux, M., Dukart, R., Maia, I. B., Kalia, S. and Schmitt-Ulms, G. (2020). Rapid Generation of Human Neuronal Cell Models Enabling Inducible Expression of Proteins-of-interest for Functional Studies. Bio-protocol 10(9): e3615. DOI: 10.21769/BioProtoc.3615.
分类
神经科学 > 发育 > 神经元
生物化学 > 蛋白质 > 荧光
细胞生物学 > 细胞工程 > CRISPR-cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link