- EN - English
- CN - 中文
Confocal and Super-resolution Imaging of RNA in Live Bacteria Using a Fluorogenic Silicon Rhodamine-binding Aptamer
含氟硅-罗丹明适配体用于活性菌中RNA的激光共聚焦超分辨率成像
发布: 2020年05月05日第10卷第9期 DOI: 10.21769/BioProtoc.3603 浏览次数: 8250
评审: Imre GáspárRosario Gomez-GarciaAnonymous reviewer(s)
Abstract
Genetically encoded light-up RNA aptamers have been shown to be promising tools for the visualization of RNAs in living cells, helping us to advance our understanding of the broad and complex life of RNA. Although a handful of light-up aptamers spanning the visible wavelength region have been developed, none of them have yet been reported to be compatible with advanced super-resolution techniques, mainly due to poor photophysical properties of their small-molecule fluorogens. Here, we describe a detailed protocol for fluorescence microscopy of mRNA in live bacteria using the recently reported fluorogenic silicon rhodamine binding aptamer (SiRA) featuring excellent photophysical properties. Notably, with SiRA, we demonstrated the first aptamer-based RNA visualization using super-resolution (STED) microscopy. This imaging method can be especially valuable for visualization of RNA in prokaryotes since the size of a bacterium is only a few times greater than the optical resolution of a conventional microscope.
Background
The visualization of specific RNA molecules by fluorescence microscopy has become invaluable during the past two decades to extend our knowledge of RNA function within cells in a spatiotemporal manner (Tyagi, 2009; Xia et al., 2017). Due to the lack of inherently fluorescent RNAs, development of fluorogenic RNA labeling tools for live-cell imaging and their adaptation to state-of-the-art microscopes–especially to super-resolution microscopes–is imperative. Super-resolution microscopy (SRM) is particularly attractive for imaging RNAs in prokaryotic systems since a bacterium is very small (~2.5 μm long, ~0.5-1 μm wide) and the resolution of a standard fluorescence microscope is restricted to ~200-300 nm due to the diffraction limit of light (Reshes et al., 2008; Nienhaus and Nienhaus, 2016). Although a plethora of tools have been developed to fluorescently label an RNA of interest (ROI), their live-cell visualization using SRM (resolution ~20-50 nm) is still challenging (Alexander and Devaraj, 2017), mainly due to the stringent requirements on the photophysical properties of the fluorophores needed for live-cell SRM. These properties include high photostability and brightness, cellular membrane permeability, water solubility, absorption and emission wavelengths preferentially in the far-red and near-infrared (NIR) region, on-off switching capability and a sufficient signal-to-background ratio during imaging (Wang et al., 2019). Thus, there is a strong need for novel RNA reporter systems that are compatible with SRM.
An especially attractive approach to visualize RNAs in live cells is the use of genetically encoded fluorescence light-up aptamers, which are short oligonucleotides that bind small, conditionally fluorescent probes with high affinity and are developed using systematic evolution of ligands by exponential enrichment (SELEX) (Holeman et al., 1998; Ouellet, 2016). Tagged to the ROI, the aptamers can be visualized due to a fluorescence increase upon small molecule binding (Figure 1). Several fluorescence enhancing RNA aptamers spanning the visible region of the spectrum have recently been developed and proven to be powerful tools for RNA labeling (Bouhedda et al., 2017; Neubacher and Hennig, 2019).
The famous Spinach, Broccoli, Mango, Corn aptamers and their improved variants bind to fluorophores with low fluorescence quantum yield in solution and become fluorescent due to the restriction of intramolecular movements upon aptamer binding (Figure 1A) (Paige et al., 2011; Filonov et al., 2014; Dolgosheina et al., 2014; Song et al., 2017). Other prominent examples are SRB-2, DNB, BHQ and Riboglow aptamers that disrupt a quencher-dye conjugate upon binding to either the dye or the quencher moiety, thereby generating fluorescence enhancement (Figure 1B) (Murata et al., 2011; Sunbul and Jäschke, 2013 and 2018; Arora et al., 2015; Braselmann et al., 2018). Most of these aptamer-ligand pairs, however, do not absorb and emit in the far-red or NIR region. Although MG, Riboglow, Mango TO-3 and the recently reported DIR-pro (which has not been reported for RNA labeling) are excited in the far-red, their fluorophores show either low photostability and brightness, phototoxicity or are hardly cell permeable (Tan et al., 2017; Autour et al., 2018; Yerramilli and Kim, 2018).
Prompted by these shortcomings, we recently developed a novel light-up RNA aptamer that binds to 3-carboxy silicon rhodamine (SiR) (Wirth et al., 2019). SiRs are an exciting new fluorophore class in the field of protein labeling for SRM because they are photostable, NIR-emitting fluorophores that change their equilibrium between the non-colored spirolactone and the fluorescent zwitterion in response to their environment (Figure 1C) (Lukinavičius et al., 2013). This property is responsible for their high cell permeability and fluorogenic behavior and contributes to the popularity of SiR dyes and their derivatives in the last decade (Wang et al., 2019). Applying SELEX on a combinatorial RNA library with a diversity of ~2 x 1015, we discovered a 50 nt-minimal aptamer, SiRA, which binds the target SiR 1 with a KD of 430 nM and shows a significant fluorescence turn-on upon dye binding (Figure 2). This finding introduces a new concept of fluorogenicity to the field of aptamer-based RNA imaging, namely environment-dependent intramolecular spirocyclization (Figure 1C). SiRA is remarkably resistant to photobleaching and constitutes the brightest far-red light up aptamer system known to date (quantum yield 0.98; extinction coefficient 86,000 M-1·cm-1). Using the SiRA system, we visualized the expression of RNAs in bacteria in no-wash live-cell imaging experiments (Wirth et al., 2019). The innovation in this protocol is that we can use SiRA’s excellent photophysical properties for stimulated emission depletion (STED) SRM of mRNA in live bacteria. To date, SiRA is the only light-up aptamer system proven to work in SRM applications.
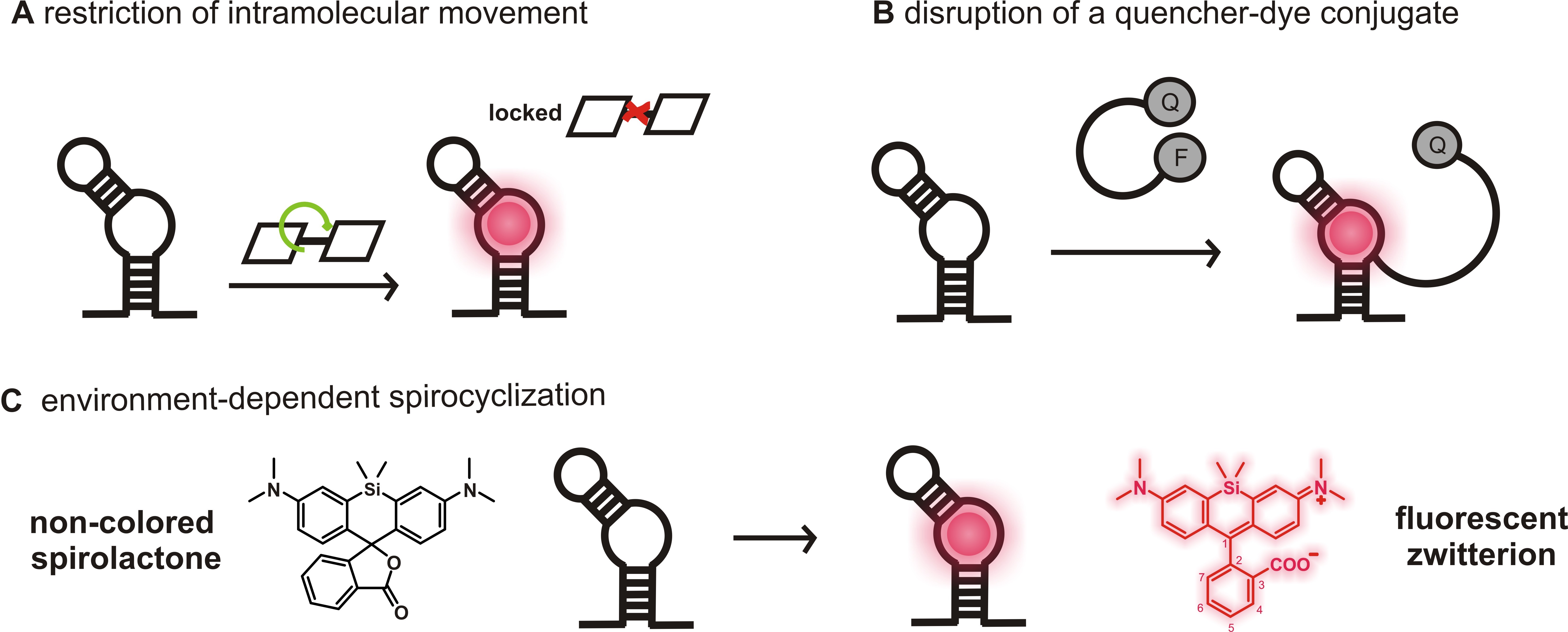
Figure 1. Fluorogenicity concepts of light-up RNA aptamers used for RNA imaging. A. A fluorophore with low fluorescence quantum yield due to molecular motions in solution (OFF) becomes highly fluorescent upon aptamer binding due to the restriction of intramolecular movements (ON). B. The contact-quenched fluorophore-quencher conjugates (OFF) light up upon binding to either the fluorophore or the quencher moiety (ON). Here, an example of a light-up system using a fluorophore binding aptamer was shown. C. In solution, SiRs can favor the non-colored spirolactone (OFF). The change in environment upon aptamer binding causes a shift of the equilibrium to the fluorescent zwitterion (ON).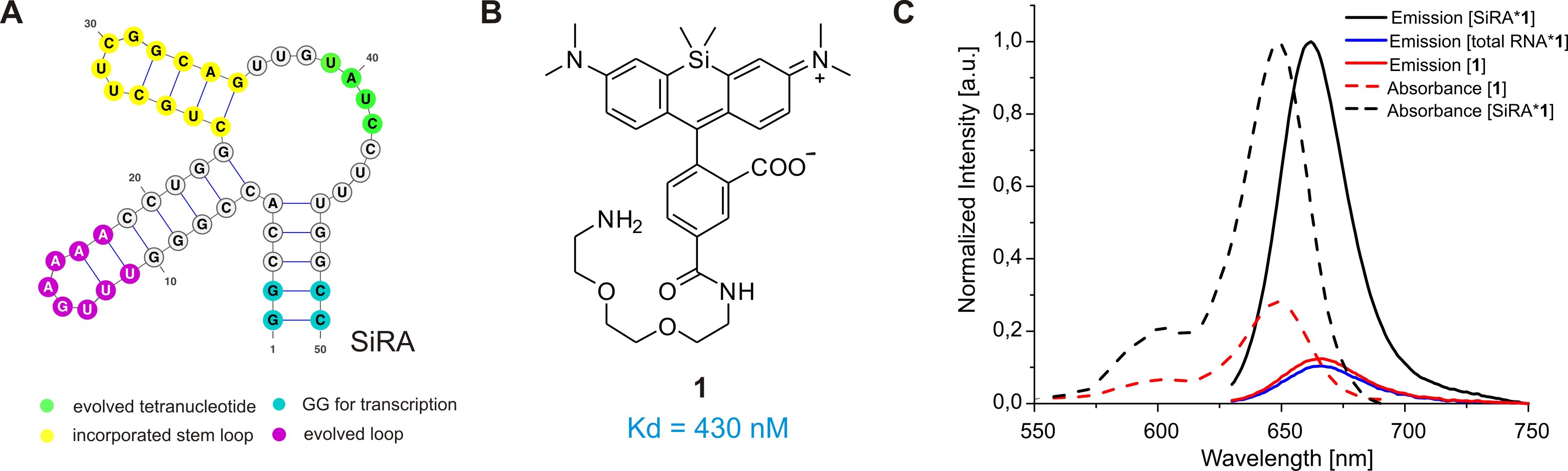
Figure 2. The SiRA aptamer and its properties. A. The predicted secondary structure of the 50-nt SiRA aptamer evolved during the selection process (important features are marked in color). B. Structure of the fluorogenic dye 1 used in this protocol, which binds to SiRA with nanomolar affinity. C. Normalized absorption and emission spectra of free SiR 1 and the [SiRA*1] complex.
In the following protocol, we describe the preparation of E. coli bacteria for live-cell confocal and STED imaging of GFP-mRNA in detail. We herein use a plasmid-based expression system and tag the GFP-mRNA with tandem repeats of the novel SiRA aptamer (SiRA5) at the 3′ untranslated region (UTR). This protocol can be adapted to visualize any plasmid expressed target RNA; however, the expression levels and the half-life of RNAs need to be kept in mind. Further optimization of the protocol might be required for specific applications.
Materials and Reagents
- Pipette tips (Sarstedt, catalog numbers: 70.113 0, 70.760.002 , 70.762 )
- Microcentrifuge tubes 1.5 ml (Sarstedt, catalog number: 72.690.001 )
- 15 μ-Slide 8-well uncoated, glass bottom #1.5 polymer coverslip (Ibidi GmbH, catalog number: 80827 )
- High clarity PP conical centrifuge tube 15 ml (Falcon®, catalog number: 352096 )
- High clarity PP conical centrifuge tube 50 ml (Falcon®, catalog number: 352070 )
- Medical Millex-GS syringe filter unit, 0.22 μm (EDM Merck, catalog number: SLGSM33SS )
- BD Discardit disposable syringes 10 ml (Becton Dickinson, catalog number: 300296 )
- PYREX® Media bottles, graduated, Corning®, 1,000 ml (VWR, PYREX®, catalog number: 1395-10L )
- FluoSpheresTM Carboxylate-Modified Microspheres, 0.04 μm, dark red fluorescent (660/680), 5% solids, azide free (Thermo Fisher Scientific, catalog number: F8789 )
- Gold beads (GC80, BB International, catalog number: EM. GC80 )
- E. coli BL21 StarTM (DE3) Invitrogen competent E. coli strains (Thermo Fisher, catalog number: C601003 ), store at -80 °C
- pET-GFP expression plasmid (pET His6 GFP TEV LIC cloning vector, 1 GFP) (Addgene, Plasmid number: 29663), store at -20 °C
- pET-GFP-SiRA5 expression plasmid, store at -20 °C
The pET-GFP-SiRA5 plasmid used in this protocol is not commercially available. Generally tandem repeats of an aptamer of choice (depending on the target RNA and expression level), can directly be fused to the ROI (in this case GFP mRNA) using proper restriction sites in the 3′ UTR (here: SalI and XhoI fast digest restriction enzymes, Thermo ScientificTM, catalog numbers: FD0644 and FD0695 , respectively) as previously described in the literature (Sunbul et al., 2018). In this study, we inserted five repeats of the SiRA aptamer with interspersed linker nucleotides according to Figure 3.
Figure 3. Excerpt of the pET-GFP-SiRA5 bacterial expression plasmid used for the visualization of GFP mRNA in this protocol. lacO: lac operator, rbs: ribosomal binding site, ROI: RNA of interest, P: promoter, T: terminator, SalI and XhoI: Restriction sites, stop: Stop codon for RNA translation.
The following sequences show an excerpt of the cloned expression vector (pET-GFP-SiRA5) as well as the pET-GFP used in this study.pET-GFP-SiRA5:black SiRA (3-nt stem loop)
...CGAGGAAACCTGTACTTCCAATCCAATATTGGAAGTGGATAACGGATCCGAATTCGAGCGCCGTCGACAGGCCCACCGGGTTTGAAAACCTGGCTGCTTCGGCAGTTGTATCCTTTGGGCCTAAGAGAGACCACCGGGTTTGAAAACCTGGCTGCTTCGGCAGTTGTATCCTTTGGTCTCAATAACCAGCCACCGGGTTTGAAAACCTGGCTGCTTCGGCAGTTGTATCCTTTGGGCCTAAGAGAGACCACCGGGTTTGAAAACCTGGCTGCTTCGGCAGTTGTATCCTTTGGTCTCAATAACCAGCCACCGGGTTTGAAAACCTGGCTGCTTCGGCAGTTGTATCCTTTGGCTGGCTCGAGCACCACCACCACCACCACTGAGATCCGGCTGCTAACAAAGCCCGAAAGGAAGCTGAGTTGGCTGCTGCCACCGCTGAGCAATAACTAGCATAACCCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTGCTGAAAGGAGGAACTATATCCGGATTGGCGAATGGGACGCGCCCTGTAGCGGCGCATTAAGCGCGGCGGGTG…
pET-GFP:
...CGAGGAAACCTGTACTTCCAATCCAATATTGGAAGTGGATAACGGATCCGAATTCGAGCGCCGTCGACAAGCTTGCGGCCGCACTCGAGCACCACCACCACCACCACTGAGATCCGGCTGCTAACAAAGCCCGAAAGGAAGCTGAGTTGGCTGCTGCCACCGCTGAGCAATAACTAGCATAACCCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTGCTGAAAGGAGGAACTATATCCGGATTGGCGAATGGGACGCGCCCTGTAGCGGCGCATTAAGCGCGGCGGGTG…
grey Variable Stem Loop or Spacer Region
blue SalI Restriction Site
pink XhoI Restriction Site - Autoclaved LB medium, Lennox (Carl Roth®, catalog number: X964.4 )
- LB Broth with agar, Lennox (Sigma-Aldrich, catalog number: L2897 )
- Kanamycin sulfate from Streptomyces kanamyceticus (Sigma-Aldrich, catalog number: K1377 ), prepare 1 M aqueous solution, filter-sterilize and store at -20 °C
- Isopropyl-β-D-thio-galactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I5502 ), prepare 1 M aqueous solution, filter-sterilize and store at -20 °C
- Potassium hydroxide pellets EMPLURA® (Sigma-Aldrich, catalog number: 1050121000 ), prepare 10 M aqueous solution
- Poly-D-lysine hydrobromide (Sigma-Aldrich, catalog number: P7886 ), prepare 2.5 mg/ml in DPBS, store at 4 °C
- Dulbecco's Phosphate Buffered Saline (DPBS) (Sigma-Aldrich, catalog number: D8537 )
- MilliQ water
- Silicon rhodamine based fluorogenic dye, 100 μM solution in DMSO
The fluorogenic dye used in this protocol is not commercially available. Therefore, it has to be synthesized and purified by HPLC as described in the literature (Wirth et al., 2019).
Note: The HPLC purification has to be carried out with great care, since the fluorescence of remaining impurities can decrease the signal-to-background ratio during the imaging experiment. - M9 Broth (Fluka Sigma-Aldrich, catalog number: 63011 )
- Magnesium sulfate monohydrate (Sigma-Aldrich, catalog number: 434183 ), prepare 1 M aqueous solution and autoclave
- D-(+)-Glucose (Sigma-Aldrich, catalog number: G8270 ), prepare 40% (w/v) aqueous solution and filter-sterilize
- Magnesium chloride hexahydrate (Sigma-Aldrich, catalog number M2670 ), prepare 1 M aqueous solution and autoclave
- Calcium chloride dihydrate (Sigma-Aldrich: catalog number: 1725801000 ), prepare 1 M aqueous solution and autoclave
- Live-cell imaging solution (Thermo Fisher Scientific, catalog number: A14291DJ )
- M9 imaging solution (see Recipes)
Equipment
- Research® plus pipettes, variable volume 200 and 1,000 μl (Sigma-Aldrich, Eppendorf®, catalog numbers: Z683817 and Z683825 )
- Steam Sterilizer (e.g., Varioklav®, model: 155S )
- Thermomixer comfort 24 x 1.5 ml (Eppendorf®)
- Incubator and shaker (Thermo Fisher Scientific, type 496)
- BioPhotometer (Eppendorf®, type 6131)
- High speed Micro Centrifuge (Neuation Technology Pvt. Ltd., catalog number: iFuge M12 )
- Freezer
- Refrigerator
- Confocal microscope (Nikon, model: A1R )
For this protocol, a point scanning confocal microscope with hybrid scanner (galvano/resonant) was used, equipped with a Nikon N Apo 60x NA 1.4 λs OI (WD 0.14 mm, FOV 0.21 x 0.21 mm) objective. For fluorescence excitation, 488 nm (green channel) and 640 nm (red channel) lasers were used. For detection, a high-sensitive GaAsP detector combined with 525/50 nm and 700/75 nm (center wavelength/bandwidth) emission filters for green and red channels, respectively, was used. - STED microscope
The STED microscope used in this protocol is home-built and has been described in detail (Gao et al., 2017). In addition, a 650-nm long-pass dichroic mirror (ET700SP; Chroma, Bellows Falls, VT) and a time-correlated single photon counting (TCSPC) card (SPC-150, Becker & Hickl GmbH, Berlin, Germany) are needed.
Note: The experiment in the protocol was carried out with 640 nm excitation and 779 nm depletion; only the vortex phase plate for plain 2D STED imaging was used.
Software
- ImageJ Fiji, version 2.0.0. with Java 1.6.0_24
- Software confocal microscope: NIS-Elements image acquisition and analysis software (Nikon)
- Software STED microscope: Imspector (Max-Planck-Innovation GmhH, Muenchen, Germany)
- Matlab (MathWorks, Natick, MA)
- OriginPro 2015 32 bit (OriginLab, Northampton, MA)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Wirth, R., Gao, P., Nienhaus, G. U., Sunbul, M. and Jäschke, A. (2020). Confocal and Super-resolution Imaging of RNA in Live Bacteria Using a Fluorogenic Silicon Rhodamine-binding Aptamer. Bio-protocol 10(9): e3603. DOI: 10.21769/BioProtoc.3603.
分类
生物化学 > RNA > RNA结构
分子生物学 > RNA > RNA 标记
分子生物学 > RNA > RNA 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link















