- EN - English
- CN - 中文
Approaching RNA-seq for Cell Line Identification
RNA序列在细胞系鉴定中的应用
发布: 2020年02月05日第10卷第3期 DOI: 10.21769/BioProtoc.3507 浏览次数: 5135
评审: Prashanth N SuravajhalaAnonymous reviewer(s)
Abstract
Cancer cell lines serve as invaluable model systems for cancer biology research and help in evaluating the efficacy of new therapeutic agents. However, cell line contamination and misidentification have become one of the most pressing problems affecting biomedical research. Available methods of cell line authentication suffer from limited access, time-consuming and often costly for many researchers, hence a new and cost-effective approach for cell line authentication is needed. In this regard, we developed a new method called CeL-ID for cell line authentication using genomic variants as a byproduct derived from RNA-seq data. CeL-ID was trained and tested on publicly available more than 900 RNA-seq dataset derived from the Cancer Cell Line Encyclopedia (CCLE) project; including most frequently used adult and pediatric cancer cell lines. We generated cell line specific variant profiles from RNA-seq data using our in-house pipeline followed by pair-wise variant profile comparison between cell lines using allele frequencies and depth of coverage values of the entire variant set. Comparative analysis of variant profiles revealed that they differ significantly from cell line to cell line whereas identical, synonymous and derivative cell lines share high variant identity and their allelic fractions are highly correlated, which is the basis of this cell line authentication protocol. Additionally, CeL-ID also includes a method to estimate the possible cross-contamination using a linear mixture model with any possible CCLE cells in case no perfect match was detected.
Keywords: Cell line authentication (细胞株鉴定)Background
Cell lines are widely used as simple model systems in biomedical research to study complex biological processes and to test the therapeutic efficacy of new agents. Its application as a model system has contributed to the characterization and understanding of important biological systems, and has made possible enormous progress in the field of biomedical research (ATCC, 2010; Yu et al., 2015), while the International Cell Line Authentication Committee (ICLAC) lists more than 400 misidentified cell lines (https://iclac.org/databases/cross-contaminations/). Contamination and misidentification of laboratory cell lines have plagued the field for more than half a century, either unintentionally or cell line changes due to passage effects, and continues to remain as one of the most prevalent problems affecting biomedical research to date. Not only it affects overall scientific reproducibility and slows down development in the field but estimated to cost billions of dollars on subsequent research work based on those irreproducible publications (Neimark, 2015; Zaaijer et al., 2017). A lot of effort has been made lately to bring attention to this widespread problem and to correct the contamination of laboratory cell lines. The impact of research on two impostor cell lines: Hep2 and INT407 was reported recently and it was estimated that $713 million were spent on the original papers published on Hep2 and INT407 and additional $3.5 billion were spent on subsequent research based on these publications (Neimark, 2015). In addition, over the last two decades, work from Dr. Korch’s group at the University of Colorado has established more than 70 commonly used cell lines, including Hep2 and INT407, to be contaminated with other cells, such as the fast-growing HeLa cancer cells (Neimark, 2015). Other reports documented that approximately 20% of cell lines currently in use are contaminated (Capes-Davis et al., 2010; Neimark, 2015; Fasterius et al., 2017) that includes many cell lines from the large datasets stored in public repositories (Fasterius et al., 2017). Thanks in part to awareness efforts towards the importance of cell line authentication many journals and institutions including NIH now require researchers to ascertain cell line integrity (Yu et al., 2015).
The most commonly used method to check the integrity of cell lines is the profiling of short tandem repeats (STRs) across several loci as recommended by the Standards Development Organization Workgroup ASN-0002 of American Type Culture Collection (ATCC) (ATCC, 2010; Yu et al., 2015; Fasterius et al., 2017; Zaaijer et al., 2017). Additionally single nucleotide polymorphism (SNP)-based assays are also used to ensure the quality and integrity of cell lines, either in combination with STR profiles or alone (Yu et al., 2015; Fasterius et al., 2017; Zaaijer et al., 2017). Both STR- and SNP-based assays have some inherent limitations. STR-based methods are not reliable in cases with microsatellite instability, loss of heterozygosity or aneuploidy as in case of many cancer cell lines, whereas currently there are no standard reference profiles available for SNP genotyping. Typically, both methods collect genomic DNA prior to the lab experiment, rather than RNAs at the beginning (control samples) as well as at the end of experiment (treated samples), most likely when cell line misuses and contaminations occur.
RNA sequencing has become a routine tool for transcriptome profiling in order to gain new biological insights. It is also being used to identify single nucleotide variants in some specific applications. Seven colorectal cell lines were re-identified by comparing their SNP based discovery profiles from RNA-seq data to the mutational profile of these cell lines available in COSMIC database (Fasterius et al., 2017). In this protocol, we present a detailed pipeline for how to avail RNA sequencing data for cell line identification. An RNA-seq based method for cell line authentication is thought to be potentially more accessible to the biomedical community as a significant number of studies involve the use of data from RNA-seq and therefore, the same can also be availed to check the integrity of the cell line without any extra reagent cost. This process includes detecting cell line specific variant profile from RNA-seq data followed by pairwise comparisons between cell lines using depth of coverage (DP) and allele frequency (FREQ) values of each variant. We have shown earlier that variant profiles are unique to each cell line and benchmarking studies including tests on independent datasets revealed that CeL-ID can identify a cell line with high accuracy (Mohammad et al., 2019). Furthermore, we also developed an additional module to identify one possible contaminator in case no perfect match was detected. In brief, we identified a total of 869,826 variants across all 934 cell lines and constructed master matrices for DP and allele frequency values where each row represents a unique variant and each column a cell line (Mohammad et al., 2019). Authentication of a query cell line includes RNA sequencing, bioinformatics analysis to identify genomic variants and subsequent comparison to DP or FREQ values of variants derived from 934 cell lines. Taken together, scientific rigor and reproducibility mandate the documentation of cell line identity and its purity before and after the biological experiment. The protocol provided utilizes the richness of RNA sequencing data to confirm the cell line integrity after each experiment.
The CeL-ID protocol can be summarized as a procedure of following steps (Figure 1). This protocol will provide details of software and program commands to complete the task.
- RNA sequencing
- Quality Assessment
- Alignment to reference genome
- Post-alignment processing
- Variant calling
- Cell line identification
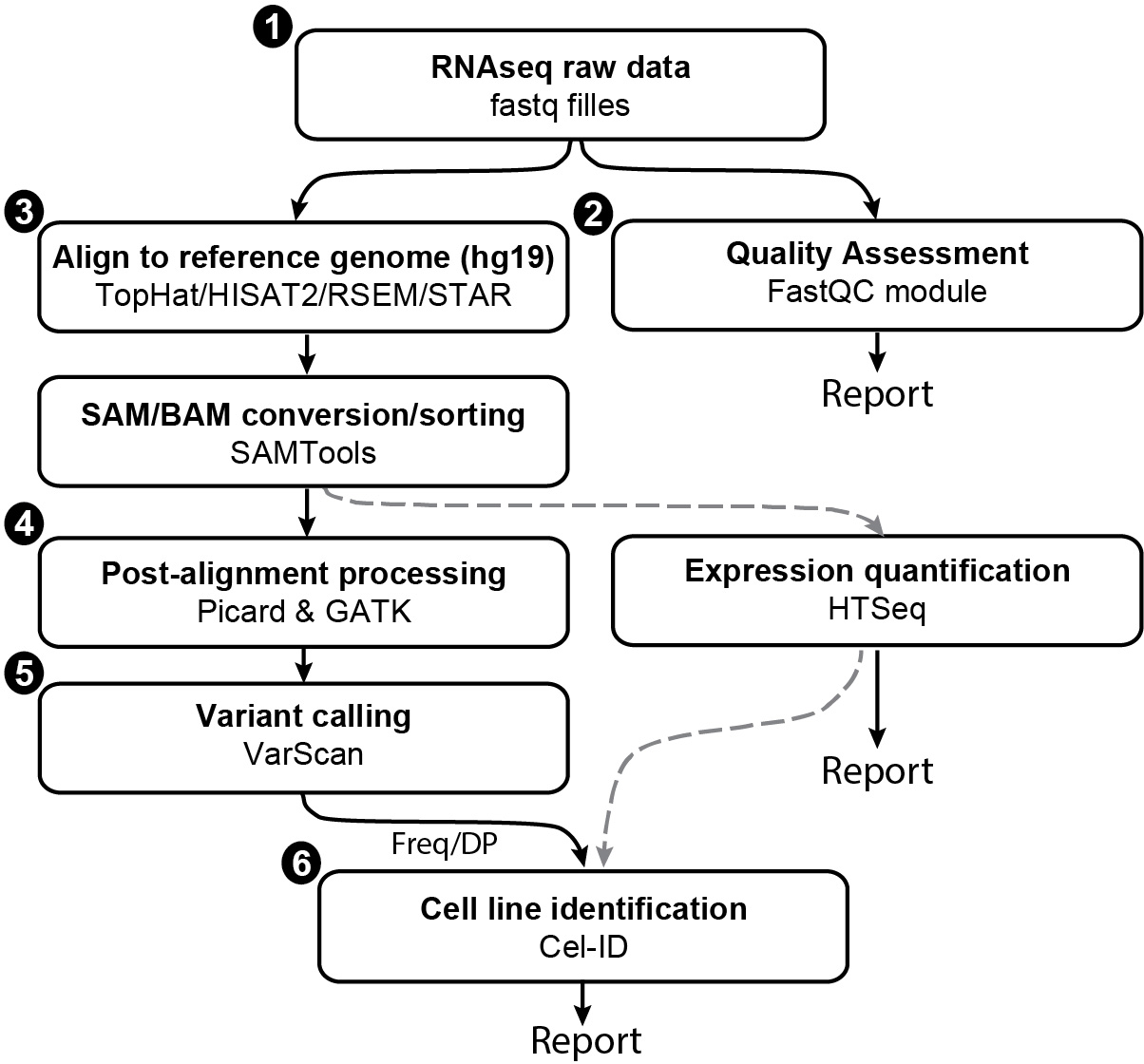
Figure 1. Flowchart showing different steps involved in cell line identification from RNA-seq data
Software
All software mentioned in this paper can be run on any workstation- or server-running UNIX based system like Linux and Mac OS. Alternatively, for Windows machines, Windows subsystem for Linux that comes with Windows 10 can be enabled to run Bash.
- FastQC v0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
- TopHat v2.1.1 (Trapnell et al., 2009) (http://ccb.jhu.edu/software/tophat/index.shtml)
- Bowtie 2 v2.3.1 (Langmead and Salzberg, 2012) (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
- HISAT2 v2.1.0 (Kim et al., 2015) (http://ccb.jhu.edu/software/hisat2/index.shtml)
- RSEM v1.2.31 (Li and Dewey, 2011) (https://github.com/deweylab/RSEM)
- STAR v2.7.3a (Dobin et al., 2013) (https://github.com/alexdobin/STAR)
- SAMtools v0.1.19 (Li et al., 2009) (http://samtools.sourceforge.net/)
- Picard v2.9.0 (https://broadinstitute.github.io/picard/)
- GATK v3.7 (DePristo et al., 2011) (https://software.broadinstitute.org/gatk/)
- VarScan v2.3.9 (Koboldt et al., 2009) (http://varscan.sourceforge.net/)
- HTSeq v0.11.1 (Anders et al., 2015) (https://htseq.readthedocs.io/en/release_0.11.1/)
All abovementioned software comes with their own README page, tutorial and/or manual detailing how to install and make use of the program. Users can choose to install software either locally or globally by installing with root/administrative privileges or just copying program executables to the /usr/bin directory (folder).
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Mohammad, T. A. and Chen, Y. (2020). Approaching RNA-seq for Cell Line Identification. Bio-protocol 10(3): e3507. DOI: 10.21769/BioProtoc.3507.
分类
癌症生物学 > 通用技术 > 遗传学 > 全基因组分析
细胞生物学 > 基于细胞的分析方法 > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link