- EN - English
- CN - 中文
A Mismatch-tolerant RT-LAMP Method for Molecular Diagnosis of Highly Variable Viruses
用于高变异病毒的分子诊断的耐错配RT-LAMP法
发布: 2019年11月05日第9卷第21期 DOI: 10.21769/BioProtoc.3415 浏览次数: 6460
评审: Alessandro DidonnaDaniela BoehmAnonymous reviewer(s)
Abstract
Loop-mediated isothermal amplification (LAMP) has been widely used in the detection of pathogens. However, there are usually numerous variants in one viral pathogen and primers employed in LAMP can hardly match all these variants. The mismatches between the primers and the viral genomes, especially those at the 3′-end of the primers, hinder LAMP reactions, leading to failure of the detection. Here, we present a mismatch-tolerant RT-LAMP protocol, which utilizes the 3′-5′ exonuclease activity of the Q5 high-fidelity DNA polymerase to remove potential mismatched bases at the 3′-end of the primers during LAMP amplification. Using HIV-1 as a proof-of-principle, we showed that this protocol could represent a promising tool for accurate detection of genetically unstable viruses in laboratory, hospital and field.
Keywords: Mismatch-tolerant RT-LAMP (耐错配的RT-LAMP法)Background
Emerging and re-emerging infectious diseases are serious threats to global public health (Mehand et al., 2018). Many outbreaks and epidemics have been caused by viruses, such as HIV-1, HCV, MERS-CoV, Ebola virus, A/H7N9 influenza virus, and more recently Zika virus. These viral diseases have led to high morbidity and mortality that disproportionally impacted on low-income countries (Van Doorn, 2017; Fenollar and Mediannikov, 2018; Waldman and Balskus, 2018). Rapid and accurate diagnosis of viral pathogens is crucial for the prevention and control of viral infectious diseases (Mehand et al., 2018).
Isothermal amplification techniques represent a promising direction for the development of point-of-care testing (POCT) diagnostic tools especially in the low-income countries or resource-limited settings (de Paz et al., 2014). Loop-mediated isothermal amplification (LAMP) is the most widely used isothermal amplification technology in biomedical research (Notomi et al., 2000). Its principle is auto-cycling strand displacement DNA amplification reaction using Bst DNA polymerase with high strand displacement activity under isothermal condition. LAMP generally uses three pairs of primers, and two inner primers (FIP and BIP) are responsible for initiating the self-primed DNA synthesis of the dumbbell form DNA. However, the biggest challenge for the detection of viruses using LAMP is the high genetic diversity of some viral genomes, which exist in the forms of genotypes, subtypes, and/or quasispecies (Sanjuan et al., 2010; Domingo and Perales, 2018). These diverse forms can easily cause mismatches with primers during their amplification, thereby resulting in a low sensitivity of detection and a limited spectrum of detection (Zhang et al., 2017; Li et al., 2019; Zhou et al., 2019). It is virtually impossible to detect all variants or serotypes in one LAMP assay as the conserved regions in the genomes are usually too short to completely match the long (approximately 40 nt) inner primers (FIP and BIP) of the assay. Therefore, an underestimate of viral load or even failure of detection is common using RT-LAMP method, especially for highly variable RNA viruses (Waldman and Balskus, 2018; Zhou et al., 2019). This may be the most important reason that limits the commercial application of LAMP in the diagnosis of viral infectious diseases (Wong et al., 2018). To overcome this problem, we recently developed a mismatch-tolerant RT-LAMP method that contains a minuscule amount of high-fidelity DNA polymerase and utilizes its 3′-5′ exonuclease activity to remove potential mismatched bases at the 3′-end of the primers during amplification (Zhou et al., 2019). The new method was demonstrated to be especially suited for the detection of highly variable viruses (Zhou et al., 2019). In this paper, we provide a detailed protocol of the mismatch-tolerant RT-LAMP method using HIV-1 detection as an example (Table 1).
Table 1. Primers used for the HIV-1 RT-LAMP assay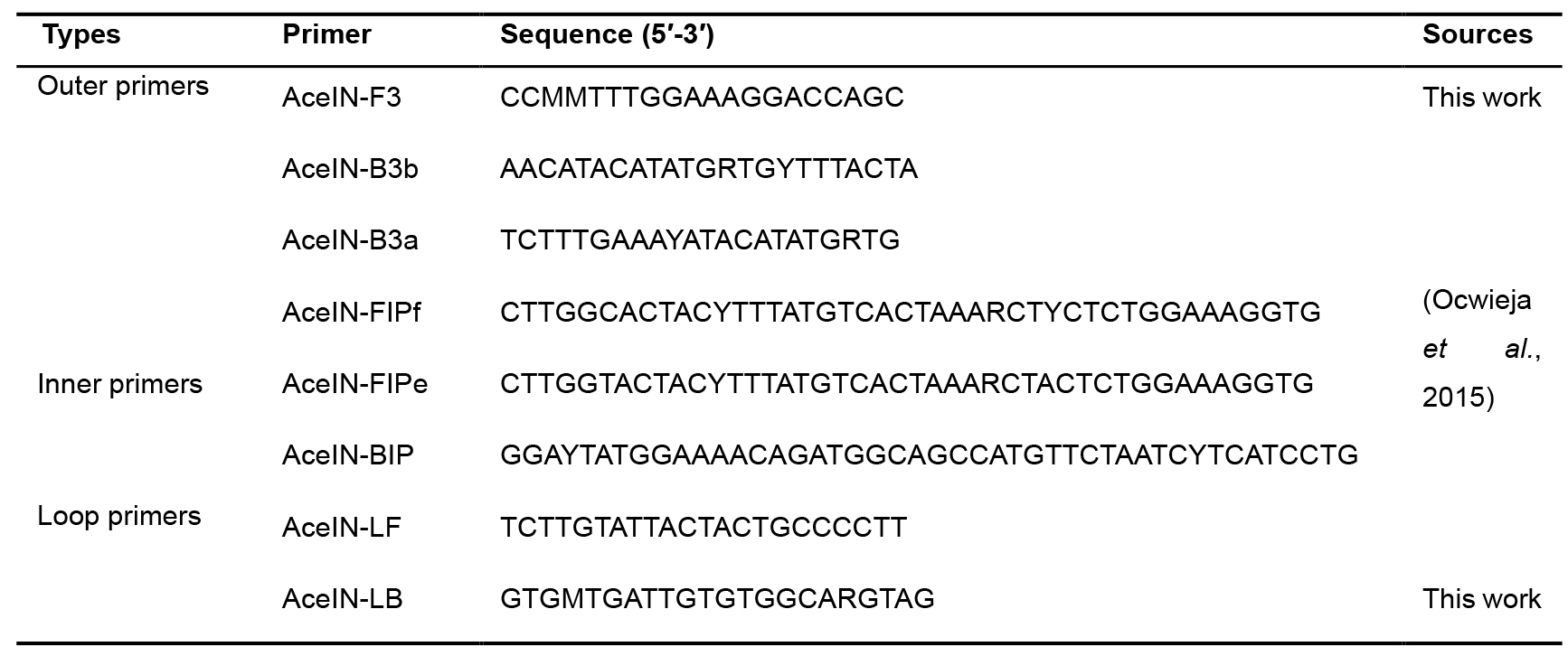
Materials and Reagents
- Axygen® MicroVolume Extended-Length Filtered Pipet Tips (Axygen, catalog number: TXLF10)
- Axygen® Universal Fit 100 μl Filtered Pipet Tips (Axygen, catalog number: TF100RS)
- Axygen® Universal Fit 200 μl Filtered Pipet Tips (Axygen, catalog number: TF200RS)
- Axygen® Universal Fit 1,000 μl Filtered Pipet Tips (Axygen, catalog number: TF1000LRS)
- Axygen 1.5 ml Snaplock Microtubes (Axygen, catalog number: MCT150CS)
- Axygen 0.2 ml PCR® Tubes (Axygen, catalog number: PCR02C)
- LightCycler® 480 Multiwell Plate 96,white (Roche, catalog number: 4729692001)
- LightCycler® 8-Tube Strips, white (Roche, catalog number: 6612601001)
- DreamTaqTM Green PCR Master Mix (2x) (Thermo Fisher Scientific, catalog number: k1081)
pUC57-IN (containing partial fragment of HIV-1 integrase gene: AF033819.3. the sequence is 5’-ACGGTTAGGGCCGCCTGTTGGTGGGCGGGAATCAAGCAGGAATTTGGAATTCCCTACAATCCCCAAAGTCAAGGAGTAGTAGAATCTATGAATAAAGAATTAAAGAAAATTATAGGACAGGTAAGAGATCAGGCTGAACATCTTAAGACAGCAGTACAAATGGCAGTATTCATCCACAATTTTAAAAGAAAAGGGGGGATTGGGGGGTACAGTGCAGGGGAAAGAATAGTAGACATAATAGCAACAGACATACAAACTAAAGAATTACAAAAACAAATTACAAAAATTCAAAATTTTCGGGTTTATTACAGGGACAGCAGAAATCCACTTTGGAAAGGACCAGCAAAGCTCCTCTGGAAAGGTGAAGGGGCAGTAGTAATACAAGATAATAGTGACATAAAAGTAGTGCCAAGAAGAAAAGCAAAGATCATTAGGGATTATGGAAAACAGATGGCAGGTGATGATTGTGTGGCAAGTAGACAGGATGAGGATTAGAACATGGAAAAGTTTAGTAAAACACCATATGTATGTTTCAGGGAAAGCTAGGGGATGGTTTTATAGACATCACTATGAAAGCCCTATCGGATCCCGGGCCCGTCGACTG-3′) (Synthesized by Shanghai BioSune Biotechnology Co., Ltd.)
- QubitTM RNA HS Assay Kit RNA (Life Technologies, catalog number: Q32855)
- WarmStart RTx Reverse Transcriptase (NEB, catalog number: M0380L)
- Q5® High-Fidelity DNA Polymerase (NEB, catalog number: M0491L)
- Bst 2.0 DNA Polymerase (NEB, catalog number: M0537L)
- Fast Mutagenesis System (Transgen, catalog number: FM111)
- QIAgen Viral RNA Mini Kit (Qiagen, catalog number: 52906)
- QIAquick® Gel Extraction Kit (Qiagen, catalog number: GC-28706)
- HiScribe T7 High Yield RNA Synthesis Kit (NEB, catalog number: E2040S)
- WarmStart® Colorimetric LAMP 2x Master Mix (DNA & RNA) (with cresol red) (NEB, catalog number: M1800S)
- Isothermal Amplification Buffer Pack (NEB, catalog number: B0537S)
- Magnesium Sulfate (MgSO4) Solution (NEB, catalog number: B1003S)
- dNTP Set, 100 mM Solutions (Thermo Fisher, catalog number: R0186)
- SYTOTM 9 Green Fluorescent Nucleic Acid Stain (Invitrogen, catalog number: S34854)
- Nuclease-Free Water (not DEPC-Treated) (AmbionTM, catalog number: AM9938)
- Agarose (Biowest, catalog number: BY-R0100)
- Ultra-pure water (Genview, catalog number: GU3313-500)
- GelRed Nucleic Acid Straining Dye (10000x) (TOROIVD, catalog number: RSD100-25)
- Plasma samples (Previous samples from our laboratory)
- Yeast extract (Oxoid, catalog number: LP0021)
- Tryptone (Oxoid, catalog number: LP0042)
- Agar (Shangxiang, catalog number: 120420)
- Sodium chloride (HuShi, catalog number: 10019318)
- Ampicillin trihydrate (Solarbio, catalog number: A7490-5)
- 50x TAE buffer (Meilunbio, catalog number: MA0004)
- 2000 DNA Marker (Yeasen, catalog number: 10501ES60)
- 5000 DNA Marker (Yeasen, catalog number: 10504ES60)
- TIANprep Mini Plasmid Kit (Tiangn, catalog number: DP103-03)
- Liquid LB medium (see Recipes)
- 100 mg/ml ampicillin solution (see Recipes)
- Ampicillin-resistant solid medium (see Recipes)
- Ampicillin-resistant liquid LB medium (see Recipes)
- 2% agarose gel (see Recipes)
- 1% agarose gel (see Recipes)
Equipment
- 0.5-10 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: I32693E)
- 10-100 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: 251596Z)
- 20-200 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: 4830359)
- 100-1,000 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: 4847859)
- NanoDropTM 2000 Spectrophotometer (Thermo Fisher, model: NanoDropTM 2000, catalog number: ND-2000)
- LightCycler® 96 System (Roche, catalog number: 05815916001)
- Bio-Rad CFX96TM Real-Time PCR System (Bio-Rad, catalog number: 785BR18555)
- Eppendorf Centrifuge 5417R (Eppendorf, model: 5417R)
- Eppendorf Mastercycler nexus (Eppendorf, catalog number:6325ZK904949)
- Tanon Gel Image System (Tanon, model: 2500)
- Tanon EPS 300 (Tanon, model: 300)
- Tanon electrophoresis tank (Tanon, model: 400)
Software
- LightCycler® 96 System software (Roche)
- Bio-Rad CFX Manager 3.1 software (Bio-Rad)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Li, Y., Zhou, Y., Ma, Y., Xu, R., Jin, X. and Zhang, C. (2019). A Mismatch-tolerant RT-LAMP Method for Molecular Diagnosis of Highly Variable Viruses. Bio-protocol 9(21): e3415. DOI: 10.21769/BioProtoc.3415.
分类
微生物学 > 病原体检测 > RT-LAMP
分子生物学 > RNA > RNA 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link














