- EN - English
- CN - 中文
Quantification of HIV-2 DNA in Whole Blood
全血HIV-2 DNA的定量分析
发布: 2019年10月20日第9卷第20期 DOI: 10.21769/BioProtoc.3404 浏览次数: 6000
评审: Alka MehraKirsten A. CoprenAswad Khadilkar
Abstract
Time to AIDS infection is longer with HIV-2, compared to HIV-1, but without antiretroviral therapy both infections will cause AIDS-related mortality. In HIV-2 infection, monitoring of antiretroviral treatment (ART) efficacy is challenging since a large proportion of HIV-2-infected individuals displays low or undetectable plasma RNA levels. Hence, quantification of cellular DNA load may constitute an alternative method for monitoring ART efficacy. Moreover, sensitive HIV-2 DNA quantification protocols are also important for the characterization of the HIV-2 reservoirs, and ultimately for the development of HIV-2 cure strategies. We have developed a sensitive and robust HIV-2 DNA quantification protocol based on whole blood as DNA source, including normalization of leukocyte cell numbers using parallel quantification of the single copy porphobilinogen deaminase gene. The specificity and sensitivity of the assay was 100%. The limit of detection was 1 copy and limit of quantification was 5 copies. When applying this protocol to HIV-2 infected, it was found that HIV-2 viral DNA was detectable in individuals in whom viral RNA was undetectable or under quantification level. Thus, this method provides a sensitive approach to HIV-2 DNA viral quantification from whole blood of HIV-2 infected patients.
Keywords: HIV-2 (HIV-2)Background
Acquired immunodeficiency syndrome (AIDS) can be caused by two viruses, either human immunodeficiency virus (HIV) type 1 (HIV-1) or type 2 (HIV-2) (Azevedo-Pereira and Santos-Costa, 2016). Although these two retroviruses have similar life cycles, they differ in that HIV-2 displays lower pathogenicity, translating into longer progression time to AIDS and reduced transmissibility (Marvin et al.,2015; Azevedo-Pereira and Santos-Costa, 2016). Another distinct feature of HIV-2 is that the plasma viral RNA load of infected individuals is significantly lower, compared with those infected with HIV-1 (Berry et al., 1998; Popper et al., 1999; Andersson et al., 2000). The cellular viral DNA load in peripheral blood has, instead, been suggested to be similar between HIV-1 and HIV-2 infections (Ariyoshi et al., 1996; Popper et al., 2000; Damond et al., 2001). However, others have indicated that also the DNA load is lower in HIV-2, compared with HIV-1, infection (Gueudin et al., 2008). Overall, HIV-2 DNA load quantification studies are relatively few, and most studies have not considered a protocol for a house-keeping gene to quantify the number of cells used in the reaction (Ariyoshi et al., 1996; Gomes et al., 1999; Popper et al., 2000; Damond et al., 2001; Soares et al., 2006; Gueudin et al., 2008; Bertine et al., 2017).
Despite the fact that the estimated time to AIDS development in HIV-2 infected individuals is longer than for individuals with HIV-1, it is clear that HIV-2 will cause AIDS-related mortality without antiretroviral therapy (ART) in infected individuals with long follow-up (Esbjornsson et al., 2018). The monitoring of treatment efficacy is, however, challenging since a large proportion of ART-naïve HIV-2 infected individuals display low or undetectable plasma RNA levels (Berry et al., 1998; Popper et al., 1999; Andersson et al., 2000; Buggert et al., 2016; Honge et al., 2018). Hence, quantification of cellular DNA load may constitute an alternative method for monitoring of ART efficacy.
For simplicity, robustness and the requirement of small volumes of blood, viral DNA quantification protocols based on whole blood, as the source of DNA, are desirable. Currently there are, however, a limited number of publications where HIV-2 DNA quantification has been based on whole blood (Bertine et al., 2017), and the need for protocols combining sensitivity with robustness is still important.
In a recent study (Esbjornsson et al., 2018), we showed that both HIV-1-infected and HIV-2-infected individuals have a high probability of developing AIDS without antiretroviral treatment.
In HIV-1 infection it has been shown that viral DNA load is an independent marker of disease progression and is well correlated with the number of latently HIV-1 infected cells, that comprises the viral reservoir (Parisi et al., 2012; Rouzioux and Avettand-Fenoel, 2018). Comprehensive studies on the HIV-2 reservoirs are, however, lacking. Thus, sensitive HIV-2 DNA quantifications protocols are also important for the characterization of the HIV-2 reservoirs, and ultimately for the development of HIV-2 cure strategies. We therefore decided to develop a sensitive and robust cellular HIV-2 DNA quantification method based on whole blood as DNA source, including normalization of leukocyte numbers using parallel quantification of the single copy porphobilinogen deaminase gene (PBDG) (Raich et al., 1986; Mbisa et al., 2009).
Materials and Reagents
- DNA extraction
- Filter tips (1,000 μl filter tip: Sarstedt, catalog number: 70.762.211; 200 μl filter tip: Sarstedt, catalog number: 70.760.211; 20 μl filter tip: Sarstedt, catalog number: 70.760.213)
- Low Binding Eppendorf tubes (DNA LoBind Snap Cap PCR Tube, 1.5 ml) (Eppendorf, catalog number: 022431021)
- PM1 cells (Lusso et al., 1995)
- Whole Blood specimen of HIV-2 infected individuals, storage in -70 °C freezer. Collect samples in EDTA tubes (BD Life Sciences, Vacutainer spray-coated K2EDTA tubes, catalog number: 367841)
- Whole Blood specimen of uninfected individuals, storage in -70 °C freezer. Collect samples in EDTA tubes (BD Life Sciences, Vacutainer spray-coated K2EDTA tubes, catalog number: 67841)
- Carrier RNA (poly A) (1350 μg) (Qiagen, catalog number: 1017647) store at room temperature before use, then store at -20 °C
- GibcoTM RPMI 1640 Medium (Thermo Fisher Scientific, Fisher Scientific, catalog number: 11875093), store at 4 °C before opening the bottle
- GibcoTM Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, Fisher Scientific, catalog number: 26140079), storage in -20 °C freezer before heat inactivation, then portion it and store at -20 °C
- GibcoTM Penicillin-Streptomycin (10,000 U/ml) (Thermo Fisher Scientific, Fisher Scientific, catalog number: 15140122), store at -20 °C
- QIAamp DNA Mini Kit (Qiagen, catalog number: 51304), store at room temperature
- QIAamp Blood DNA Mini Kit (Qiagen, catalog number: 51104), store at room temperature
- GibcoTM Phosphate buffered saline (PBS, pH = 7.4) (Thermo Scientific, Fisher Scientific catalog number: 10010023), store at room temperature
- Complete RPMI-1640 medium (see Recipes)
- Preparation of Standards
- Filter tips (1,000 μl filter tip: Sarstedt, catalog number: 70.762.211; 200 μl filter tip: Sarstedt, catalog number: 70.760.211; 20 μl filter tip: Sarstedt, catalog number: 70.760.213)
- Pipette Tips (1,000 μl filter tips: Sarstedt, catalog number: 70.762.100; 200 μl filter tips: Sarstedt, catalog number: 70.760.502; 10 μl tips: Sarstedt, catalog number: 70.1130)
- Low Binding Eppendorf tubes (DNA LoBind Snap Cap PCR Tube, 1.5 ml) (Eppendorf, catalog number: 022431021)
- MicroAmpTM Optical 8-Tube Strip, 0.2 ml (Thermo Scientific, Applied BiosystemsTM, catalog number: 4316567)
- MicroAmpTM Optical 8-Cap Strips (Thermo Scientific, Applied BiosystemsTM, catalog number: 4323032)
- HIV-2 (NIH-Z Strain) Purified Virus and electron microscopy counted (Advanced Biotechnologies Inc, catalog number: 10-127-000), storage in -70 °C freezer
- Qiagen miRNeasy micro Kit (Qiagen, catalog number: 217084), store at 4 °C
- DNA oligonucleotides primers were obtained from Invitrogen and listed in Table 1, store oligonucleotides at -20 °C
- SuperScriptTM III One-Step RT-PCR System with PlatinumTM Taq DNA Polymerase (Thermo Fisher Scientific, Invitrogen, catalog number: 12574018), storage in -20 °C freezer. SuperScriptTM III One-Step RT-PCR System is supplied with the following:
- SuperScript® III RT/Platinum® Taq Mix (50 μl)
- 2x Reaction Mix (containing 0.4 mM of each dNTP, 3.2 mM MgSO4) (1 ml)
- 5 mM Magnesium Sulfate (500 μl)
- PlatinumTM Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific, Invitrogen, catalog number: 11304011), storage in -20 °C freezer. Platinum® Taq DNA Polymerase High Fidelity (20 μl, at 5 U/μl) is supplied with the following:
- 10x High Fidelity Buffer [600 mM Tris-SO4 (pH 8.9), 180 mM (NH4)2SO4] (1.25 ml)
- 50 mM MgSO4 (1 ml)
- Agarose (Thermo Fisher Scientific, Invitrogen, catalog number: 16500500), store at room temperature
- Milli-Q quality water (RNase, DNase free water [dH2O]) (VWR, catalog number: SH30538.02), store at room temperature
- AccuGENETM 50x TAE Buffer (Lonza, catalog number: 51216), store at room temperature
- Gel Red (VWR International, catalog number: 41003), store at room temperature
- QIAquick PCR purification Kit (Qiagen, catalog number: 28104), store at room temperature
Table 1. Primers used for HIV and PBDG standard preparation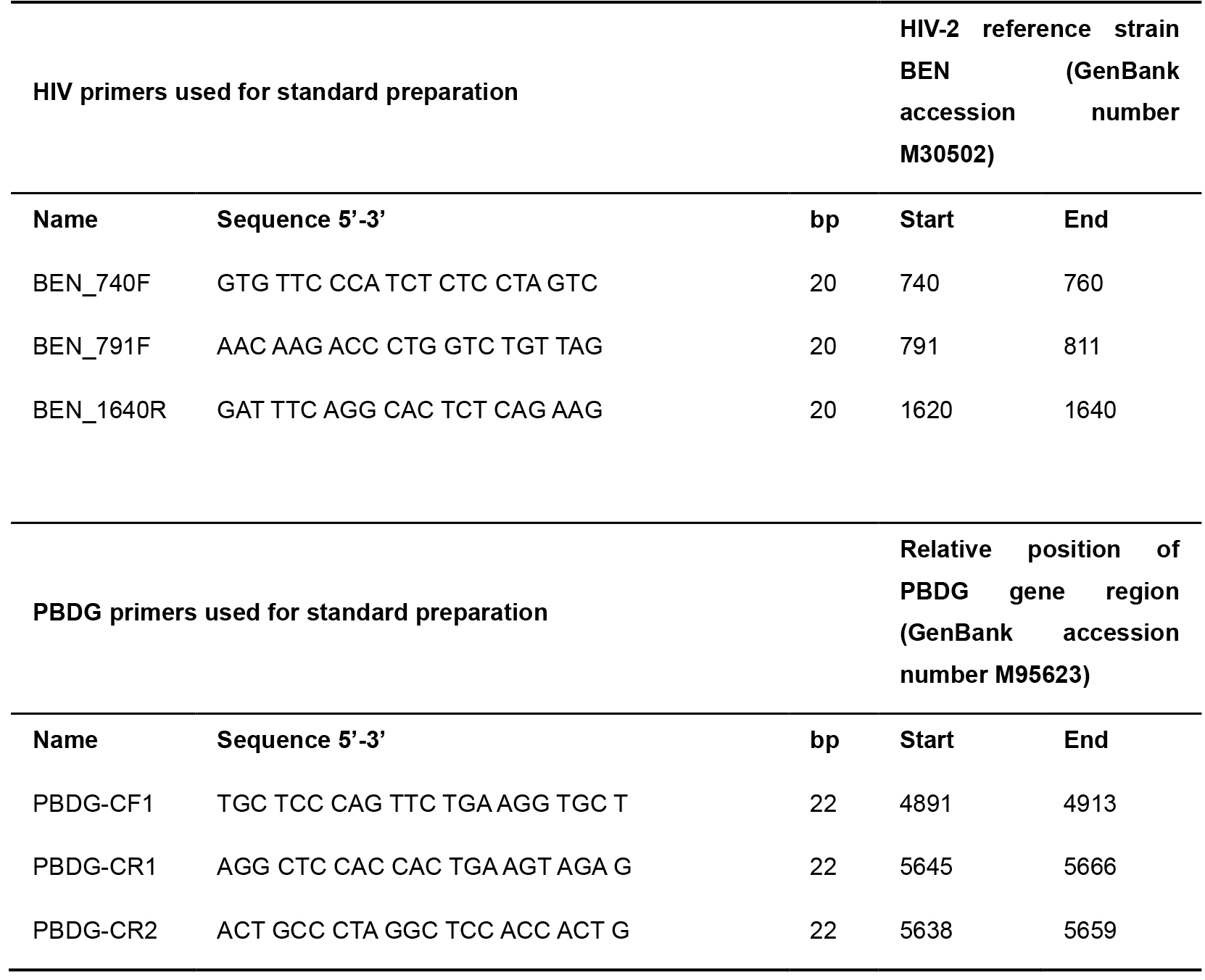
- qPCR for HIV-2 DNA detection
- Pipette tips (1,000 μl filter tips: Sarstedt, catalog number: 70.762.100; 200 μl filter tips: Sarstedt, catalog number: 70.760.502)
- 96-well qPCR plates (Applied BiosystemsTM, catalog number: 4346906)
- Optical Adhesive Film (Applied BiosystemsTM, catalog number: 4360954)
- HyCloneTM Water, Molecular Biology Grade (RNase, DNase, proteinase free) (Thermo Fisher Scientific, Fisher Scientific, GE Healthcare HyClone SH30538.FS, catalog number: 10247783), store at room temperature
- HIV-1 infected U937 Cells (U1) (Folks et al., 1987)
- Maxima probe qPCR Master Mix (2x) (Thermo Scientific, catalog number: K0231), storage in -20 °C freezer. qPCR Master Mix contains the following:
- 2x Maxima Probe/ROX qPCR Master Mix containing Maxima Hot Start Taq DNA Polymerase and dNTPs (1.25 ml)
- Nuclease-Free Water (1.25 ml)
- Primer and probe sequences for HIV-2 was obtained from Damond et al. (2002) (PMID: 12354861). Storage in -20 °C freezer
- Primer and probe sequences of the housekeeping human gene PBGD was obtained from Mbisa et al. (2009) (PMID 19020818). Storage in -20 °C freezer
- Agarose (Thermo Fisher Scientific, Invitrogen, catalog number: 16500500), store at room temperature
- Milli-Q quality water (RNase, DNase free water [dH2O]) (VWR, catalog number: SH30538.02), store at room temperature
- AccuGENETM 50x TAE Buffer (Lonza, catalog number: 51216), store at room temperature
- Gel Red (VWR International, catalog number: 41003), store at room temperature
- 1 Kb Plus DNA Ladder (Thermo Fisher Scientific, Invitrogen, catalog number: 10787018), storage in -20 °C freezer
Equipment
- EVETM Automated Cell Counter, NanoEnTek (VWR, Avantor, catalog number: 10027-452)
- EVETM Cell Counting Slides with Trypan Blue (VWR, Avantor, catalog number: 10027-446)
- Eppendorf Microcentrifuge, model: 5415D, with rotor F 45-24-11, AC/DC input 230 V AC, 50-60 Hz (Sigma-Aldrich, catalog number: Z604062)
- Life Eco Thermal Cycler, 96 well gradient, 100-240V, 50-60Hz, 600W, (Bioer Technology Co., catalog number: BTC42096)
- Gilson single-channel/adjustable-volume pipettes: 0.1-2 μl, 2-20 μl, 20-200 μl, 100-1,000 μl (Eppendorf, catalog number: F167380)
- Applied Biosystems StepOnePlus Real-Time PCR System (Thermo Scientific, Applied BiosystemsTM, catalog number: 4376600)
- NanoDrop ND-1000 spectrophotometer (Thermo Scientific, catalog number: ND-2000C)
- Mini ReadySub-CellTM electrophoresis system (Bio-Rad Laboratories, catalog number: 1704467)
- UV-Transparent gel tray (Bio-Rad Laboratories, catalog number: 1704435)
- 8-well Comb (Bio-Rad Laboratories, catalog number: 1704463)
- Gel DocTM XR+ Gel Documentation System (Bio-Rad Laboratories, catalog number: 1708195)
Software
- Image Analysis Software, Image LabTM Software (Life Sciences, Bio-Rad)
- Excel 2016 (Microsoft)
- GraphPad Prism 7.0.
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Szojka, Z., Karlson, S., Jansson, M. and Medstrand, P. (2019). Quantification of HIV-2 DNA in Whole Blood. Bio-protocol 9(20): e3404. DOI: 10.21769/BioProtoc.3404.
分类
微生物学 > 微生物遗传学 > DNA > DNA 定量
分子生物学 > DNA > DNA 定量
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link