- 提交稿件
- 订阅
- CN
- EN - English
- CN - 中文
- EN - English
- CN - 中文
Immunoprecipitation and Sequencing of Acetylated RNA
乙酰化RNA的免疫沉淀和测序
发布: 2019年06月20日第9卷第12期 DOI: 10.21769/BioProtoc.3278 浏览次数: 6987
评审: Gal HaimovichVaibhav B. ShahAnca Flavia Savulescu

相关实验方案
基于适体的 mRNA 亲和纯化程序 (RaPID) 用于鉴定酵母中的相关 RNA (RaPID-seq) 和蛋白质 (RaPID-MS)
Rohini R. Nair [...] Jeffrey E. Gerst
2022年01月05日 3948 阅读
Abstract
Generation of the epitranscriptome through chemical modifications of protein-coding messenger RNAs (mRNAs) has emerged as a new mechanism of post-transcriptional gene regulation. While most mRNA modifications are methylation events, a single acetylated ribonucleoside has been described in eukaryotes, occurring at the N4-position of cytidine (N4-acetylcytidine or ac4C). Using a combination of antibody-based enrichment of acetylated regions and deep sequencing, we recently reported ac4C as a novel mRNA modification that is catalyzed by the N-acetyltransferase enzyme NAT10. In this protocol, we describe in detail the procedures to identify acetylated mRNA regions transcriptome-wide using acetylated RNA immunoprecipitation and sequencing (acRIP-seq).
Keywords: N4-acetylcytidine (N-乙酰胞嘧啶)Background
Chemical modifications of RNA have emerged as a new mechanism for the post-transcriptional regulation of gene expression (Zhao et al., 2017). In contrast to a handful of well-studied modified bases that constitute the DNA epigenome, over a hundred modified ribonucleosides are present in all types of RNA. This diversity offers new possibilities to modulate gene expression, significantly expanding the metabolic and regulatory functions of RNA (Zhao et al., 2017). Mainly studied in abundant transfer RNA (tRNA) and ribosomal RNA (rRNA), ribonucleoside modifications have more recently been described in messenger RNAs (mRNAs), where they form the basis of the epitranscriptome (Zhao et al., 2017). These epitranscriptomic modifications have the potential to control all steps of mRNA metabolism including structure, stability, location and translation.
Several modifications, including N6-methyladenosine (m6A), N6,2’-O-dimethyladenosine (m6Am), N1-methyladenosine (m1A), pseudouridine (Ψ), inosine (I), 5-methylcytidine (m5C), 5-hydroxylmethylcytidine (hm5C), 7-methylguanosine (m7G) and 2’-O-methylnucleosides (Nm) are found in eukaryotic mRNA and can influence the metabolism and function of mRNA (Li et al., 2016). While the majority of mRNA modifications are methylation events, only a single acetylated ribonucleoside has been described in eukaryotes, occurring at the N4-position of cytidine (N4-acetylcytidine, ac4C, Figure 1A). ac4C is one of the universally conserved RNA modifications present in all domains of life and had previously been observed in rRNA and tRNA (Ito et al., 2014a and 2014b; Sharma et al., 2015). More recently, ac4C was observed in RNA viruses and in polyadenylated RNA (poly(A)-RNA) of yeast and human cancer cells (Dong et al., 2016; McIntyre et al., 2018; Tardu et al., 2018), raising the possibility that ac4C is present in mRNA. In all cases, ac4C production was attributed to the enzyme NAT10 (Figure 1A), or its homologs (Ito et al., 2014a and 2014b; Sharma et al., 2015), suggesting a non-redundant activity.
To investigate the occurrence of ac4C in human mRNA, we generated tools for antibody-based enrichment and transcriptome-wide mapping, a method we named acRIP-seq (Arango et al., 2018). Additionally, to assess ac4C regulation and function, we deleted NAT10 in HeLa cells. The sum of these studies confirmed NAT10-dependent acetylation of mRNAs (Arango et al., 2018). Overall, acetylation was enriched within coding sequences and loss of ac4C through NAT10 ablation led to a specific decrease in the expression of defined ac4C targets. More specifically, we found that ac4C increases mRNA stability and enhances translation efficiency of mRNAs when the modification is present in coding sequences (Arango et al., 2018).
Here, we report a detailed protocol for acetylated RNA immunoprecipitation and sequencing, acRIP-seq (Figures 1B-1C). This method is used to identify acetylated mRNA regions transcriptome-wide.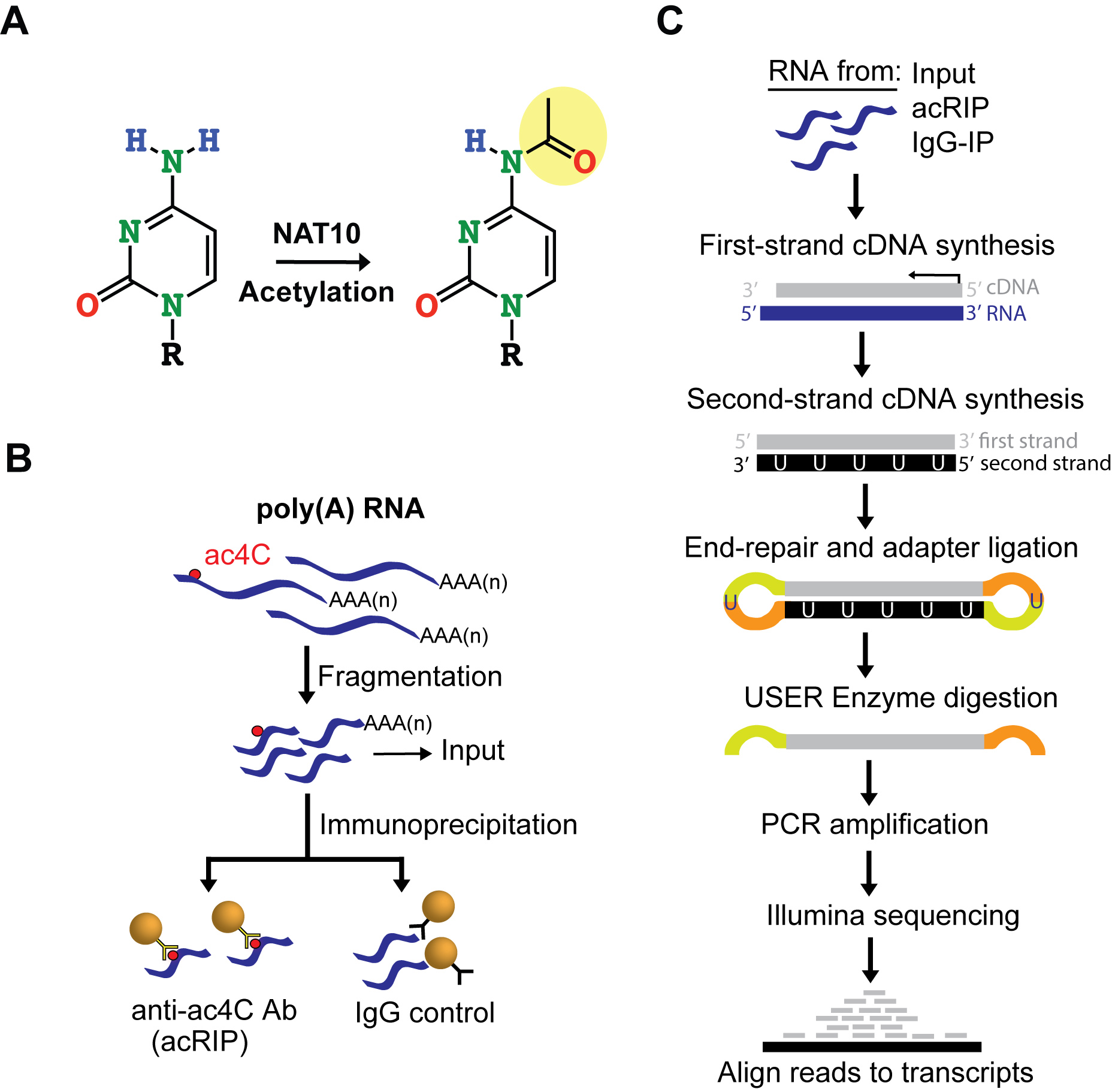
Figure 1. acRIP-seq identifies acetylated regions within mRNA transcriptome-wide. A. NAT10 catalyzes cytidine acetylation. B-C. Schematics of acRIP-seq. Fragmented poly(A) RNA is immunoprecipitated (IP) with anti-ac4C antibodies or Isotypic IgG control (B). RNA from acRIP, IgG-IP and Inputs are used to construct Illumina sequences using the NEBNext® UltraTM II Directional RNA Library protocol followed by high-throughput sequencing and bioinformatical analysis to identify putative acetylated regions transcriptome-wide (C). USER: Uracil-Specific Excision Reagent.
Materials and Reagents
- Pipette tips (RNase-free)
- 0.2 ml PCR tubes (RNase-free)
- 1.5 ml microcentrifuge tubes (RNase-free)
- 2.0 ml microcentrifuge tubes (RNase-free)
- NEBNext® Magnesium RNA Fragmentation Module (NEB, catalog number: E6150)
- NEBNext® UltraTM II Directional RNA Library Prep Kit (NEB, catalog number: E7760S)
- NEBNext® Multiplex Oligos for Illumina® (Index Primers Set 1, NEB, catalog number: E7335S)
- RNase inhibitor, murine (NEB, catalog number: M0314S)
- Agilent RNA 6000 kit (Agilent, catalog number: 5067-1511)
- Agencourt AMPure XP Beads (Beckman Coulter, catalog number: A63881)
- Agencourt RNAclean XP Beads (Beckman Coulter, catalog number: A63987)
- MAXIscript T7 Transcription Kit (Thermo Fisher Scientific, catalog number: AM1312)
- Protein G-Magnetic Dynabeads (Thermo Fisher Scientific, catalog number:10003D)
- DynabeadsTM oligo(dT)25 (Thermo Fisher Scientific catalog number: 61002)
- Linear acrylamide (Thermo Fisher Scientific, catalog number: AM9520)
- Acid-Phenol:Chloroform, pH 4.5 (Thermo Fisher Scientific, catalog number: AM9722)
- Proteinase K, RNase-free (Thermo Fisher Scientific, catalog number: AM2546)
- EDTA, RNase-free (Thermo Fisher Scientific, catalog number: AM9260G)
- Sodium Acetate, RNase-free (Thermo Fisher Scientific, catalog number: AM9740)
- Nuclease-free H2O (Thermo Fisher Scientific, catalog number: AM9938)
- Tris-HCl, RNase-free (Thermo Fisher Scientific, catalog number: 15567027)
- NaCl, RNase-free (Thermo Fisher Scientific, catalog number: AM9759)
- SDS, RNase-free (Thermo Fisher Scientific, catalog number: AM9823)
- Bovine Serum Albumin (BSA, Thermo Fisher Scientific, catalog number: AM2616)
- Ethanol (Fisher Scientific, catalog number: BP2818100)
- Chloroform (Fisher Scientific, catalog number: BP1145-1)
- Sodium Phosphate monobasic (Sigma-Aldrich, catalog number: S0751)
- Sodium Phosphate dibasic (Sigma-Aldrich, catalog number: S3264)
- Triton X-100 (Sigma-Aldrich, catalog number: 93443)
- Rabbit monoclonal IgG isotype control (Cell Signaling Technologies, catalog number: 3900S)
- Rabbit monoclonal anti-N4-acetylcytidine (Abcam, catalog number: ab252215)
- ac4CTP (Trilink, catalog number: C-0001)
- Purified poly(A) RNA
- Acetylated RNA (see section C: In vitro transcription of acetylated RNA)
- LiCl (Sigma-Aldrich, catalog number: L7026)
- 10x acRIP buffer (see Recipes)
- Proteinase K buffer (see Recipes)
- Poly(A) RNA binding buffer (see Recipes)
- Poly(A) RNA washing buffer (see Recipes)
Equipment
- Pipette 0.2 to 2 μl (Thermo Fisher Scientific, catalog number: 4642010)
- Pipette 2 to 20 μl (Thermo Fisher Scientific, catalog number: 4642060)
- Pipette 20 to 200 μl (Thermo Fisher Scientific, catalog number: 4642080)
- Pipette 100 to 1000 μl (Thermo Fisher Scientific, catalog number: 4642090)
- -20 °C freezer (General Electric, model FUM 21SVCRWW)
- -80 °C freezer (Thermo Fisher Scientific, model Ultima Plus)
- Thermal Cycler (Thermo Fisher Scientific, catalog number: 4484073)
- ThermoMixer (Eppendorf, catalog number: 2231000574)
- Refrigerated Microcentrifuge (Eppendorf, catalog number: 5404000138)
- Rotator RotoFlex (Denville Scientific, catalog number: H5700)
- Magnetic rack for 1.5 ml tubes DynaMagTM-2 Magnet (Thermo Fisher Scientific, catalog number: 12321D)
- Magnetic rack for 0.2 ml tubes DiaMag02 (Diagenode, catalog number: B04000001)
- Nanodrop Spectrophotometer (Nanodrop, catalog number: ND-1000)
- Agilent 2100 Bioanalyzer (Agilent, catalog number: G293BA)
- HiSeq 2500 Illumina sequencer (Illumina, model HiSeq 2500)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Arango, D., Sturgill, D. and Oberdoerffer, S. (2019). Immunoprecipitation and Sequencing of Acetylated RNA. Bio-protocol 9(12): e3278. DOI: 10.21769/BioProtoc.3278.
分类
分子生物学 > RNA > RNA 纯化
分子生物学 > RNA > 表观转录组
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
提问指南
+ 问题描述
写下详细的问题描述,包括所有有助于他人回答您问题的信息(例如实验过程、条件和相关图像等)。
Share
Bluesky
X
Copy link