- EN - English
- CN - 中文
Visualizing Filamentous Actin in Chlamydomonas reinhardtii
在莱茵衣藻中利用鬼笔环肽进行纤维状肌动蛋白可视化分析
发布: 2019年06月20日第9卷第12期 DOI: 10.21769/BioProtoc.3274 浏览次数: 6010
评审: Samantha E. R. DundonKae-Jiun ChangAnonymous reviewer(s)
Abstract
This protocol aims to visualize the filamentous actin network in Chlamydomonas reinhardtii. We improved fixed-cell labeling conditions using the F-actin probe, phalloidin. We created a Chlamydomonas-optimized protocol by halving the phalloidin incubation time, electing for optimal fixation conditions, and selecting for a healthy cell population. This phalloidin protocol is quick, effective, and is the only labeling method to date that allows for reliable actin filament detection in fixed vegetative Chlamydomonas cells. This method reveals previously unidentified actin structures in Chlamydomonas and novel insights into cytoskeletal dynamics.
Keywords: Chlamydomonas reinhardtii (莱茵衣藻)Background
Chlamydomonas reinhardtii is a single-celled green alga widely used in studying photosynthesis, cilia, chloroplast biology, and the synthesis of biofuels. It is a leading model system in these fields due to the organism’s well-characterized two apical flagella, inexpensive and simple culture conditions, and a fully sequenced haploid genome allowing for excellent genetic studies. Understanding the Chlamydomonas cytoskeleton will play an important role in advancing these areas of research. However, the filamentous actin network of Chlamydomonas reinhardtii remains uncharacterized, partly due to the difficulties in visualizing filaments in this eukaryotic alga.
Recently, there has been some success in detecting filamentous actin in Chlamydomonas. Through the expression of the fluorescently-tagged actin binding peptide, LifeAct, live-cell imaging revealed an actin-based perinuclear structure (Avasthi et al., 2014; Onishi et al., 2016). However, there is increasing evidence that LifeAct alters cell morphology, actin organization, actin-dependent functions, and preferentially labels a subset of actin structures (Courtemanche et al., 2016, Flores et al., 2019). Further, achieving stable expression is difficult in Chlamydomonas due to epigenetic silencing of exogenous genes (Cerutti et al., 1997) and the need to colocalize actin probes with fixed cell organelle markers is essential for understanding the revitalized field of Chlamydomonas actin biology. Thus, an efficient and specific fixed-cell method of actin visualization would lead to significant advancement in the field and offer novel insights into basic actin biology.
Available actin antibodies do not discriminate between monomeric and filamentous actin. It was previously shown that the widely used fluorescent actin probe phalloidin was ineffective in labeling filamentous actin in vegetative Chlamydomonas cells (Harper et al., 1992). However, phalloidin does label an actin-dense structure in gametic cells known as the fertilization tubule (Detmers et al., 1985). By optimizing phalloidin staining conditions, we are now able to visualize the Chlamydomonas actin network with exquisite detail in vegetative cells (Figure 1). Specifically, reducing the staining incubation time from 30 min to 16 min was essential in achieving optimal signal to noise which allowed for detection above Chlamydomonas’s auto-fluorescent chloroplast. Usage of a brighter and more photostable fluorophore, Atto 488, was also critical in obtaining a high signal to noise ratio and uniform labeling. This protocol allows us to understand how cell body actin filaments are distributed in gametic and vegetative cells for the first time. The previously unidentified filamentous actin structures will be crucial in exploring actin-dependent behaviors and how co-expressed actin genes in Chlamydomonas function to regulate cytoskeletal dynamics. Development of these strategies may prove useful for the study of actin filaments in other protists for which actin visualization has also been challenging.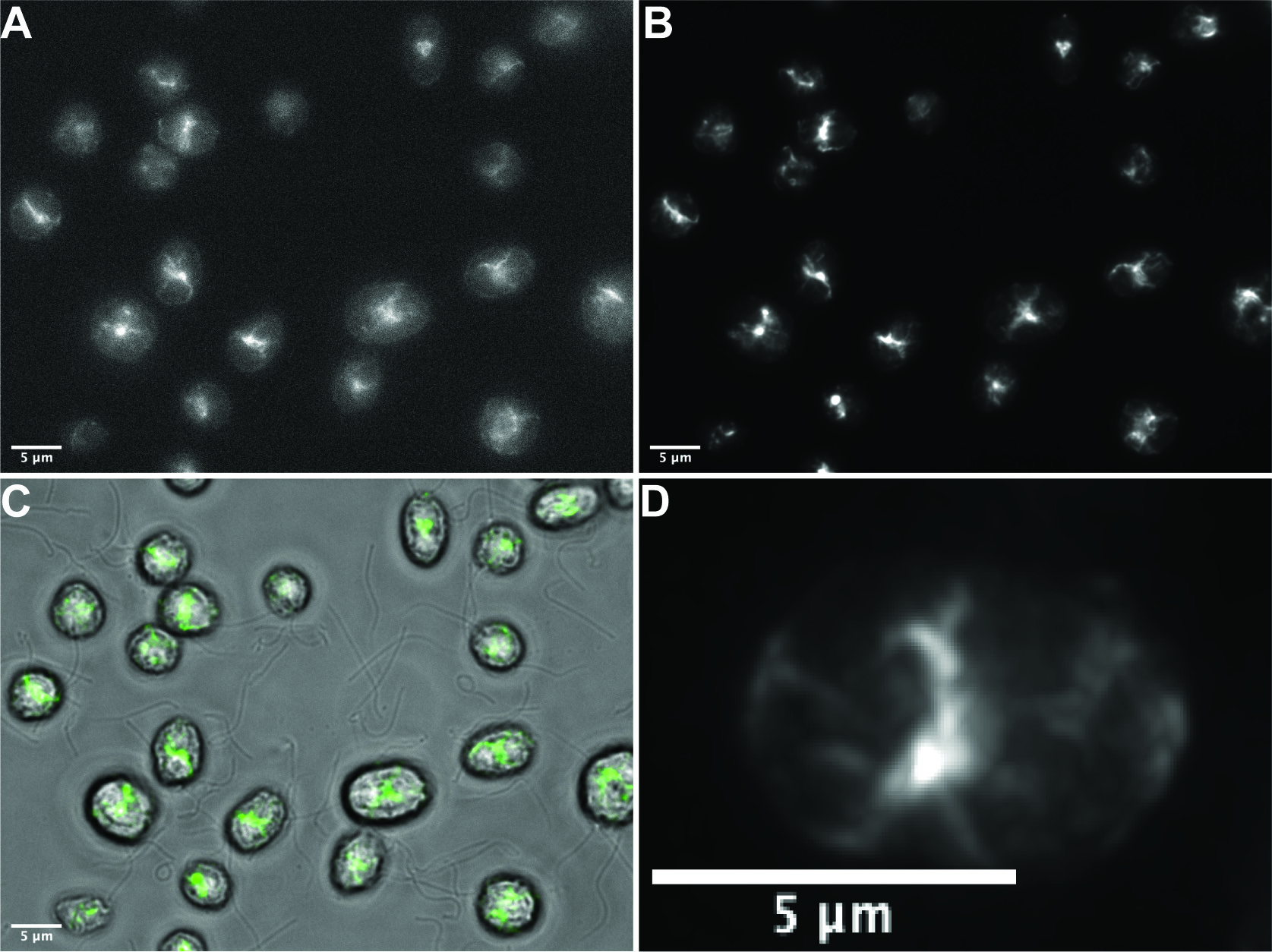
Figure 1. Phalloidin staining in wild-type vegetative Chlamydomonas reinhardtii. A. Representative raw single widefield image of specific filamentous actin staining using our optimized protocol. B. The deconvolved maximum intensity projection of a Z-stack (0.3 μm steps) of the field visualized in A. C. Overlay of brightfield and fluorescence channels with phalloidin signal indicated in green. D. Deconvolved maximum intensity projection of a Z-stack (0.3 μm steps) of a single cell demonstrating effective phalloidin staining. Scale bars = 5 μm.
Materials and Reagents
- Kimwipes
- Parafilm
- Hydrophobic Marker (Elite PAP Pen, Diagnostic Biosystems Ref: K039)
- 1.5 ml Microcentrifuge Tubes
- Columbia Jars (DWK Life Sciences WheatonTM Columbia Jars for Coverslips, catalog number: 02-912-637)
- 10 μl inoculating loop
- Culture tubes (Borosilicate Glass Tubes 13 x 100 mm item) (Globe Scientific, catalog number: 1510)
- Micropipette tips
- Microscope slides
- Coverglass (22 x 22 mm square, 0.13 mm to 0.16 mm thick) (Corning, catalog number: 2845-22)
- Disposable Petri Dishes 60 x 15 mm (VWR®, catalog number: 25384-164)
- Whatman® Qualitative Filter Paper, Grade 1 (Whatman, catalog number: 1001 125)
- Chlamydomonas Strains
Note: The wild-type strain CC-125 mating type + was obtained from the Chlamydomonas Resource Center (University of Minnesota). - Atto 488 Phalloidin (Sigma, catalog number: 49409), store at -20 °C, shield from light
- Poly-L-lysine solution (Sigma, catalog number: P8920)
- PBS Tablet (Research Products International PBS 100 ml Tablets SKU P32080-100T)
- PFA aqueous solution 16% (Electron Microscopy Sciences, catalog number: 15710)
- HEPES (Corning, catalog number: 61-034-RM)
- NaOH pellets (J.T. Baker, catalog number: 3722)
- HCl (Fischer Chemical, catalog number: A481-212)
- Acetone (Histological) (Fisher Chemical, catalog number: A16P-4)
- Fluoromount-GTM (Invitrogen, Ref: 00-4958-02)
- DI Water
- BD Biosciences DifcoTM Agar, Granulated (Ref: 214530)
- Hunter’s Trace Elements (Can be ordered from the Chlamydomonas Resource Center)
- Acetic Acid, Glacial (Certified ACS) (Fisher Chemical, catalog number: A35S-500)
- Methanol
- Tris Base (C4H11NO3), Ultra Pure
- Ammonium Chloride (NH4Cl)
- Magnesium Sulfate Heptahydrate (MgSO4·7H2O)
- Calcium Chloride Dihydrate (CaCl2·2H2O)
- Potassium Phosphate Dibasic Anhydrous (K2HPO4)
- Potassium Phosphate Monobasic (KH2PO4)
- 1x PBS (see Recipes)
- 4% PFA in 7.5 mM HEPES Buffer (see Recipes)
- Phalloidin Stock Preparation (see Recipes)
- TAP liquid media (Tris Acetate Phosphate Media) (see Recipes)
- 100x Tris buffer (see Recipes)
- 100x TAP Salts (see Recipes)
- 1000x Phosphate Solution (see Recipes)
- 1.5% TAP Agar Plates (see Recipes)
Equipment
- Chemical spatula
- Graduated cylinder
- Beaker
- Stir plate
- Stir rod
- pH meter
- Scale
- Tweezers
- Pipettes
- Gusto® High-Speed Mini Centrifuge (Heathrow Scientific, catalog number: HEA10050)
- Empty glove boxes, cover to shield samples from light
- Microscope for image acquisition (We use a Nikon Eclipse Ti-S equipped with a QiIMAGING QICAM)
- Roller Drum (Cel Gro Tissue Culture Rotator) (Thermo Scientific, catalog number: 1640Q)
- -20 °C freezer
Software
- Huygens Essential (Scientific Volume Imaging)
- ImageJ (Schneider et al., 2012)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Craig, E. W. and Avasthi, P. (2019). Visualizing Filamentous Actin in Chlamydomonas reinhardtii. Bio-protocol 9(12): e3274. DOI: 10.21769/BioProtoc.3274.
分类
细胞生物学 > 细胞染色 > 蛋白质
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link